Look at this movie. The young chemist that you see at the beginning and at the end was born after 2D NMR. You'd expect she's at ease with it. The video is instead about a new product devoted to the semi-automated analysis of 1D spectra only. What happened? The guys at ACD Labs have recycled a few pieces of their code and created a new attractive product, called "1D NMR Assistant". It's encouraging to see new ideas in the field of NMR processing (although the ingredients aren't new, the overall concept is). There's another movie into Ryan's blog and others are coming. They save me the effort to actually try the program itself. I will cite a few sentences from Ryan's.
ACD/1D NMR Processor...can sometimes be viewed as bloated, and overly complex for a novice or non-expert user.
You are saying it! And you haven't even mentioned the 2D Processor! The 1D processor is already bloated by itself, according to you.
From the very beginning, we were developing a product with the NMR Spectroscopist in mind.
I can't buy it. From the very beginning the name of the company was "Advanced Chemistry Development", not "Advanced Spectroscopy Development". If you really had the spectroscopist in mind, why you removed 2D?
We have just de-emphasized some of those features in the software in an effort to greatly simplify the toolbars and interface. I believe that we reduced the learning curve significantly.
Admirable effort. It's a relief to hear somebody else recognizing what I am repeating from a lifetime: too many icons don't make an easier-to-use program.
The ultimate motivation behind this product was the simple idea that, "an NMR spectrum is a means to an end"
What's the end? Please don't conclude that the report is the end (the movie doesn't say such a thing). Anyway: the assistant can create the report from the spectrum even when the chemist understands the bare minimum.
There is nothing available that really helps users evaluate and interpret an NMR spectrum and it's relationship with a chemical structure. That is until now, IMHO. I think the development of ACD/1D NMR Assistant changes all that.
It changes that, but not all that. In the movie all the multiplets are isolated.
In my practice this is a rare circumstance, but evidently my compounds were more complicated than the average.
This product wasn't just built by a few software-oriented people within ACD/Labs.
We already know you are a chemist.
Over the next few weeks, I will be highlighting different features and workflows in the software to educate you on how it works and what it does.
This leaves me the burden to educate you on what it does not.
The assistant assumes that any multiplet can be analyzed regardless of the rest of the spectrum. It's the so-called "first order approximation". The lower the magnetic field, the less frequently this approximation can be applied. What happens in the remaining cases? If you recognize that there is strong coupling, either you resort to second-order analysis or you renounce to extract both the coupling constants and the chemical sifts (reporting a range for the latter). If you don't recognize it, the extracted parameters will not be accurate.
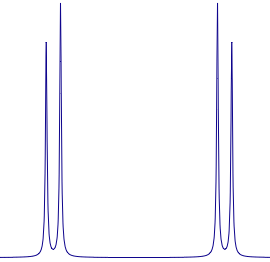 A spectrum subjected to strong coupling is not necessarily complicated. Look at this familiar AB pattern. Everybody knows the rule: the J corresponds to the splitting, while the chemical shift corresponds to center of mass of the doublet (not to the average position!).
A spectrum subjected to strong coupling is not necessarily complicated. Look at this familiar AB pattern. Everybody knows the rule: the J corresponds to the splitting, while the chemical shift corresponds to center of mass of the doublet (not to the average position!).Another celebrated case is represented by the ODCB. Here you really need to perform a generalized simulation, taking all signals into account. These cases are rare indeed, but if you are serious about extracting NMR parameters you can't avoid quantum mechanic forever.
 In one of the videos (the first one the blog, Nov 15) the assistant fails, at first, to extract the Js from a triplet of triplets. Then Ryan manually peak-picks two shoulders and feeds them to the program, which eventually succeeds. That's great, but can you always do without a line-fitting deconvolution? What's more, the example highlights the importance of the visual inspection. In the old days of CW NMR, the spectrum was plotted on a large sheet, similar to the A3 format (as we call it here in Europe). When I look at the ACD reports, where the actual plot is reduced to little more than a thumbnail, I wonder: "Do they observe them with a magnifying glass?". Of course the user is free to print magnified plots and this is not a terrible limitation, but what about the rendering on the screen? A few months ago Ryan admitted that his software was not as good-looking as the alternatives on the market, from which I concluded, erroneously, that the next version of his software would have filled the gap. No way, the graphics are still aliased, and, believe me, it was a little painful to see those movies. It brings you a decade back. When you get used to anti-aliased graphics, like in my case, you can't go back. Here is the aliased version of my previous ODCB picture.
In one of the videos (the first one the blog, Nov 15) the assistant fails, at first, to extract the Js from a triplet of triplets. Then Ryan manually peak-picks two shoulders and feeds them to the program, which eventually succeeds. That's great, but can you always do without a line-fitting deconvolution? What's more, the example highlights the importance of the visual inspection. In the old days of CW NMR, the spectrum was plotted on a large sheet, similar to the A3 format (as we call it here in Europe). When I look at the ACD reports, where the actual plot is reduced to little more than a thumbnail, I wonder: "Do they observe them with a magnifying glass?". Of course the user is free to print magnified plots and this is not a terrible limitation, but what about the rendering on the screen? A few months ago Ryan admitted that his software was not as good-looking as the alternatives on the market, from which I concluded, erroneously, that the next version of his software would have filled the gap. No way, the graphics are still aliased, and, believe me, it was a little painful to see those movies. It brings you a decade back. When you get used to anti-aliased graphics, like in my case, you can't go back. Here is the aliased version of my previous ODCB picture. It contains a good measure of shoulders, and they are all artifacts. While it's normally possible to discriminate authentic shoulders from fake ones like these, through a torture for your eyes, today we have eliminated the fake shoulders altogether. Anti-aliasing is implemented in hardware, not in software. Your graphic card is already capable to draw as shown in the first two examples. Those more advanced applications that draw better are doing nothing more than allowing the graphic cards to do their work. ACD Labs found the time to reassemble their old routines into a new attractive package but did not find the time to bring their drawing routines into the third millennium. What a pity!
It contains a good measure of shoulders, and they are all artifacts. While it's normally possible to discriminate authentic shoulders from fake ones like these, through a torture for your eyes, today we have eliminated the fake shoulders altogether. Anti-aliasing is implemented in hardware, not in software. Your graphic card is already capable to draw as shown in the first two examples. Those more advanced applications that draw better are doing nothing more than allowing the graphic cards to do their work. ACD Labs found the time to reassemble their old routines into a new attractive package but did not find the time to bring their drawing routines into the third millennium. What a pity!Do you know what else is missing? 2D spectroscopy. Maybe next year they'll be offering you the 2D NMR assistant...