Requires Mac OS 10.3, is Universal binary, reads Varian, Bruker, Jeol, JCAMP-DX, Tecmag, SwaN-MR, Siemens, GE medical, Spinsight files; can process, fit and simulate; creates pictures and PDF files; price: 30 euro (includes unlimited updates).
Friday, 15 December 2006
iNMR reader
Requires Mac OS 10.3, is Universal binary, reads Varian, Bruker, Jeol, JCAMP-DX, Tecmag, SwaN-MR, Siemens, GE medical, Spinsight files; can process, fit and simulate; creates pictures and PDF files; price: 30 euro (includes unlimited updates).
Thursday, 14 December 2006
Freeware
A well written list of NMR freeware can be found at:
http://www.pharmacy.umaryland.edu/PSC/NMR/proc_anal/tecmag.html
It's 3 years old, which is, unfortunately, too much for such a rapidly evolving field. Apart from this inconvenient, it's nearly perfect. The only (minor) defect is that the list contains more than one entry per operative system. My ideal list would just contain a unique recommendation per OS.
The questions is: why there is so much NMR freeware around, that all web lists are rapidly outdated? It is well proven that the authors of freeware never become famous, and I mean any kind of freeware, I don't mean the NMR niche. If NMR freeware is never celebrated, and if the author of any free program is never remembered, the probability of becoming famous writing it are zero. Only one word can explain why people writes it: money. They write it for the money and they stop writing it when they don't receive money anymore. This has been my case and the case of all the NMR freeware I know about. I used to receive my regular salary when writing freeware and renounced to it to write commercial software.
Unfortunately, for the users, the day arrived when the sponsors realized that this kind of software brings no fame. The only hope is to become the standard of the field. NMRPipe went near to that status, yet was limited in platforms (only UNIX) and in applications (mainly 2D). All that the sponsor can get is a publication, written by the author himself. In some cases they may add a poster or a follow-up paper, and by that time they realize they are scraping the bottom of the barrel. No external author ever cares to write a review. Even youtube movies get their reviews, but NMR softwares don't. Vomiting as it can be, this is the reality. When the sponsor realizes that even the stinkiest movie gets more attention than the best-crafted NMR program, that's the moment when the flow of money stops. And when the money arrives no more the programmer (being him no saint) quits.
I don't mean that it would have been different, but it certainly wouldn't have hurt, if the beneficiaries had cited more frequently their softwares, reviewd them, evangelized them. The beneficiaries of freeware also receive their salaries and their grants, therefore their choice was not dictated by economical motivations. If they had really loved the free software they'd a least tried to save it. They didn't love and didn't care. They decided for the death of NMR freeware and, if it was not for the residual stupidity of the sponsors, they'd had almost fulfilled their intent.
How a software dies? It is constantly fighting on four fronts:
The lethal bullet can arrive from any of these sources, but the first cause alone would suffice. NMR is so vast a field that all programs have at least one hole: they never find the time to cover the whole gamma.
At the risk of being liable, today I uploaded the definitive version of SwaN-MR at http://www.inmr.net/index.html#DOWN. The sponsor forgot its existence. I wrote it but it's not mine. If you are so poor that you couldn't buy a new Mac in the last four years, chances are that SwaN-MR will run on your old machine. If you are so lucky to own a vintage 68K Mac (mine died), look somewhere else.
http://www.pharmacy.umaryland.edu/PSC/NMR/proc_anal/tecmag.html
It's 3 years old, which is, unfortunately, too much for such a rapidly evolving field. Apart from this inconvenient, it's nearly perfect. The only (minor) defect is that the list contains more than one entry per operative system. My ideal list would just contain a unique recommendation per OS.
The questions is: why there is so much NMR freeware around, that all web lists are rapidly outdated? It is well proven that the authors of freeware never become famous, and I mean any kind of freeware, I don't mean the NMR niche. If NMR freeware is never celebrated, and if the author of any free program is never remembered, the probability of becoming famous writing it are zero. Only one word can explain why people writes it: money. They write it for the money and they stop writing it when they don't receive money anymore. This has been my case and the case of all the NMR freeware I know about. I used to receive my regular salary when writing freeware and renounced to it to write commercial software.
Unfortunately, for the users, the day arrived when the sponsors realized that this kind of software brings no fame. The only hope is to become the standard of the field. NMRPipe went near to that status, yet was limited in platforms (only UNIX) and in applications (mainly 2D). All that the sponsor can get is a publication, written by the author himself. In some cases they may add a poster or a follow-up paper, and by that time they realize they are scraping the bottom of the barrel. No external author ever cares to write a review. Even youtube movies get their reviews, but NMR softwares don't. Vomiting as it can be, this is the reality. When the sponsor realizes that even the stinkiest movie gets more attention than the best-crafted NMR program, that's the moment when the flow of money stops. And when the money arrives no more the programmer (being him no saint) quits.
I don't mean that it would have been different, but it certainly wouldn't have hurt, if the beneficiaries had cited more frequently their softwares, reviewd them, evangelized them. The beneficiaries of freeware also receive their salaries and their grants, therefore their choice was not dictated by economical motivations. If they had really loved the free software they'd a least tried to save it. They didn't love and didn't care. They decided for the death of NMR freeware and, if it was not for the residual stupidity of the sponsors, they'd had almost fulfilled their intent.
How a software dies? It is constantly fighting on four fronts:
- advancements in NMR
- changes in file formats
- changes in the operative systems, processors, compilers, libraries, etc...
- competition from other NMR programs
The lethal bullet can arrive from any of these sources, but the first cause alone would suffice. NMR is so vast a field that all programs have at least one hole: they never find the time to cover the whole gamma.
At the risk of being liable, today I uploaded the definitive version of SwaN-MR at http://www.inmr.net/index.html#DOWN. The sponsor forgot its existence. I wrote it but it's not mine. If you are so poor that you couldn't buy a new Mac in the last four years, chances are that SwaN-MR will run on your old machine. If you are so lucky to own a vintage 68K Mac (mine died), look somewhere else.
Tuesday, 12 December 2006
XWin-NMR
For the most part of its life-cycle, XWin-NMR did contain a huge hole and nobody used to complain. I wonder if Bruker is a cult more than a instruments-maker. Whatever Bruker does is forgiven by its loyal customers. In the case I am writing about, you had a value of temperature stored in your file that had nothing to do with the actual temperature at which the spectrum was acquired. For example: the probe was at 240 K and the software reported the value 298 K. The reason was obvious, almost natural: the temperature could be controlled by Win-NMR, but in practice you were (and still are) using another program, called "edte", which operates directly on the thermal unit and can't be aware of the active data-set, or of the existence of a spectrometer, for that matter. With edte you can set and monitor the temperature of the probe, but the value is not saved in the spectrum. (The latter belonging to XWin-NMR, not to edte).
It was unbelievable that at the same time Bruker could claim to care about GLPs. The situation went on, in this fashion, for many years. The last generation of XWin-NMR solved the problem: when the acquisition is completed, the software reads the temperature of the probe and stores this value into the spectrum.
This anecdote tells why old Bruker users admire the successor TopSpin. The cause of their happiness is the end of their past sufferings.
It was unbelievable that at the same time Bruker could claim to care about GLPs. The situation went on, in this fashion, for many years. The last generation of XWin-NMR solved the problem: when the acquisition is completed, the software reads the temperature of the probe and stores this value into the spectrum.
This anecdote tells why old Bruker users admire the successor TopSpin. The cause of their happiness is the end of their past sufferings.
Monday, 11 December 2006
Nuts
I ignore the history of Acorn and generally don't like to express my thoughts about people. Unfortunately I assigned to myself the task of defeating the generalized laziness of the web and now I am forced to describe Acorn's main product, called NUTS (NMR Utility Transform Software). The bad thing is not NUTS, but the laziness of the web. You can find countless lists of NMR software whose only effect is a great loss of time for all their readers. Most of the programs you can find have serious limitations and can be considered unusable. The compilers of the list never find the time to download, test the programs, write a review and discourage you from doing the same.
This particular case (NUTS) is a borderline one. The program is worth to see because is a living fossil, just like its nice vintage icon suggests. Using NUTS can be as funny as visiting a museum. The web site is informative, readable, moderately vintage (by today's web standards) and certainly a valuable source of information. I appreciate a lot the simplicity of the pricing policy, its transparency, the immediate availability of a downloadable demo, etc.. a lot of things that I am in tune with.
There is nothing wrong in writing a program in an ancient DOS-like style. It's the right answer to the hundreds of icons of Topspin (there are no icons into Nuts). I am seriously concerned, however, at the mere idea that in 2006 a chemist could invest money in buying such a fossil. The user can only open a single window with a single pane and two spectra cannot remain open simultaneously. There is something more unusual, though. In a single word: this program is modal, if you know what it means. Before testing Nuts I believed that all Bruker software was modal. Today I must say that Bruker software is 40% modal, while NUTS is pure modal. It's so modal that the manual doesn't even mention the existence of modes and calls them "subroutines". There are 9 of them, plus the starting mode, called "base level". In simpler words: there is a total of 10 different interfaces (just like ten different programs) and the user is expected to learn them all. I wonder how somebody can remember where the commands are because, even when the menus bear familiar names like "Edit" and "View", the menu items are highly non-standard. It's not a matter of opinion, is a matter of design. I mean: if you are writing a program for the Macintosh, like Acorn is doing, you should adhere to clearly stated principles and practices. The principle is that half of your commands should be the familiar "Copy", "Paste", "Undo"... in their familiar menu positions.
The good thing is that you can just ignore the menus. All the NUTS command can and should be written in the command line. Menus have an aesthetic function. In the basic level you find six equivalent menu commands: "Processing Parameters", "Set LB", "Apply EM", "Apply GM", "Set Sine Phase", "Apply Sine Multiply". They all open the same dialog window!
A noteworthy property of most menu items is that they are not context sensitive. They are never dimmed, even when there is no spectrum "on stage". In practice, half of the commands, if given at the start of the program, crash the program itself! It's also funny to see that they make no distinction of FID or spectrum. Knowing "what's on" is 100% responsibility of the user. The sine-bell multiplication, for example, that is intended for FIDs, can easily be applied to transformed spectra, even if I can't find no good reason for such an operation.
All the above things are today considered "non-professional", yet they represent surmountable defects. The average user can discriminate by himself if he is working on a FID or on a spectrum and he will soon learn to do without menus and use the command line exclusively. Now you also know why there is no "Undo" command: the program crashes as soon as you make a mistake, nobody had ever the opportunity of using "Undo" and it was eventually removed. Using the same logic, however, most of the remaining menu items could have removed: For example, the first 6 items of the "Process" menu, plus the six other items already cited, could all have been put into a single dialog.
In my exploration I haven't found the commands to enhance/decrease the apparent intensity of the spectrum. It may be a limitation of the demo version, but at this point I am full of doubts and believe in nothing. Is NUTS actually "usable"? The web site says that they are developing the Mac version because they received specific requests in that direction. How is it possible that somebody who loves the Mac can order a program driven by the command line? How is it possible that they sell NUTS 1D at 499 USD and the fully fledged Mestre-C sells at 265 euro only?
This review refers to wxNuts version 0.6.0, compiled on October 19, 2006.
This particular case (NUTS) is a borderline one. The program is worth to see because is a living fossil, just like its nice vintage icon suggests. Using NUTS can be as funny as visiting a museum. The web site is informative, readable, moderately vintage (by today's web standards) and certainly a valuable source of information. I appreciate a lot the simplicity of the pricing policy, its transparency, the immediate availability of a downloadable demo, etc.. a lot of things that I am in tune with.
There is nothing wrong in writing a program in an ancient DOS-like style. It's the right answer to the hundreds of icons of Topspin (there are no icons into Nuts). I am seriously concerned, however, at the mere idea that in 2006 a chemist could invest money in buying such a fossil. The user can only open a single window with a single pane and two spectra cannot remain open simultaneously. There is something more unusual, though. In a single word: this program is modal, if you know what it means. Before testing Nuts I believed that all Bruker software was modal. Today I must say that Bruker software is 40% modal, while NUTS is pure modal. It's so modal that the manual doesn't even mention the existence of modes and calls them "subroutines". There are 9 of them, plus the starting mode, called "base level". In simpler words: there is a total of 10 different interfaces (just like ten different programs) and the user is expected to learn them all. I wonder how somebody can remember where the commands are because, even when the menus bear familiar names like "Edit" and "View", the menu items are highly non-standard. It's not a matter of opinion, is a matter of design. I mean: if you are writing a program for the Macintosh, like Acorn is doing, you should adhere to clearly stated principles and practices. The principle is that half of your commands should be the familiar "Copy", "Paste", "Undo"... in their familiar menu positions.
The good thing is that you can just ignore the menus. All the NUTS command can and should be written in the command line. Menus have an aesthetic function. In the basic level you find six equivalent menu commands: "Processing Parameters", "Set LB", "Apply EM", "Apply GM", "Set Sine Phase", "Apply Sine Multiply". They all open the same dialog window!
A noteworthy property of most menu items is that they are not context sensitive. They are never dimmed, even when there is no spectrum "on stage". In practice, half of the commands, if given at the start of the program, crash the program itself! It's also funny to see that they make no distinction of FID or spectrum. Knowing "what's on" is 100% responsibility of the user. The sine-bell multiplication, for example, that is intended for FIDs, can easily be applied to transformed spectra, even if I can't find no good reason for such an operation.
All the above things are today considered "non-professional", yet they represent surmountable defects. The average user can discriminate by himself if he is working on a FID or on a spectrum and he will soon learn to do without menus and use the command line exclusively. Now you also know why there is no "Undo" command: the program crashes as soon as you make a mistake, nobody had ever the opportunity of using "Undo" and it was eventually removed. Using the same logic, however, most of the remaining menu items could have removed: For example, the first 6 items of the "Process" menu, plus the six other items already cited, could all have been put into a single dialog.
In my exploration I haven't found the commands to enhance/decrease the apparent intensity of the spectrum. It may be a limitation of the demo version, but at this point I am full of doubts and believe in nothing. Is NUTS actually "usable"? The web site says that they are developing the Mac version because they received specific requests in that direction. How is it possible that somebody who loves the Mac can order a program driven by the command line? How is it possible that they sell NUTS 1D at 499 USD and the fully fledged Mestre-C sells at 265 euro only?
This review refers to wxNuts version 0.6.0, compiled on October 19, 2006.
Topspin NMR free download
Did you know there is a long list of NMR programs that can be freely downloaded from the internet and work with your Bruker files? Check them out!
iNMR reader
Spinworks
MatNMR
NPNMR
NPK
CCPN
Other free programs are available upon request (with some restrictions):
Jeol Delta
NMRPipe
NMRnotebook
ACD
Nuts
iNMR reader
Spinworks
MatNMR
NPNMR
NPK
CCPN
Other free programs are available upon request (with some restrictions):
Jeol Delta
NMRPipe
NMRnotebook
ACD
Nuts
Saturday, 9 December 2006
NMR googling

Building your personal NMR database couldn't be simpler. All you have to do is to process your spectra as always. They are automatically indexed by Spotlight and can be searched either with Spotlight or with Speclight (free).
It has always been possible, on spectrometer of any brand, to store a "title" that describes the sample and the experiment. iNMR automatically imports that text and feeds Spotlight with it and with other notes you insert with the acclaimed iNMR annotation tools. To increase your searching options you can also:
- Include the elemental formula of your compounds (and of your impurities too!). If you include this information you can perform searches based on the molecular weight, the number of carbon atoms, etc...
- Include the SMILES strings of your compounds (it's a simple matter of Copying & Pasting). If you include this information you can perform searches based on molecular fragments.
- Perform peak-picking. If you are not used to it, start today. Even if you hide the output, you'll be able to find peaks wherever they are, simply typing a range of chemical shifts. Very handy to recognize impurities!
All your spectra will be indexed, old and new ones, after you install iNMR 1.6. You can also use Speclight to retrieve chemical files (indexed by ChemSpotlight).
Sunday, 3 December 2006
Speclight

Speclight is a free application, with apparently no precedent, that retrieves NMR spectra and other documents in the file system (even on a network). Requires Mac OS X Tiger and, possibly, iNMR 1.6. When I say "other documents" I mean those indexed by ChemSpotlight. Both the cost and the effort of building a personal NMR database are suddenly reduced to zero. Speclight will debut next week; source code available upon request.
Tuesday, 28 November 2006
I did it!
Friday, 24 November 2006
Pepper
Mac users are usually proud of their machines and they should be even more so. Their operative system has an hidden engine running, very quietly, and putting everything into a catalog. It's called Spotlight and it's not secret at all, because every Tiger owner has used it at least once. In practice it's a substitute for the human memory. You type a word like "pepper" and it searches your computer for everything related to it: recipes, pictures, mails, pdf documents and, last but not least, the music of the late great Art Pepper. It works like Google but it's even faster: the first results appear before you type the final "r" in pepper! This engine can, potentially, search anything. You can, for example ask: "Search on my computer all carbon spectra containing a peak between 90 and 95 ppm" and can also restrict the search to a specific magnetic field, or solvent, or year. Potentially you can also search for all molecules containing a given substructure. The great thing is that it all comes with no effort. It's not like creating and building a database. The system does everything automatically every time a file is copied, changed or deleted. And it's also free. Do you want to learn how to do it?
You understand that I will be busy in the next few weeks...
- The first component is the the terminal command "mdfind". The simplest example of usage is: "mdfind pepper", which is equivalent to typing "pepper" into the Spotlight search field. Substitute a more articolate query expression for "pepper" and you can modulate the search anyway you like.
- A Spotlight plug-in that can parse NMR documents. The forthcoming version of iNMR (1.6) comes with a bundled plugin. A plugin can also be a stand-alone bundle. It is possible to create plugins for every kind of documents, generated by any application. For example you can create a plugin for Varian spectra, copy all your back-ups on the hard disc, and have all them indexed.
- Possibly a graphic interface that hides the complexity of the terminal to the average Mac user. Just like the plug-in, it can be written by any volunteer, it must not necessarily reside inside an NMR application. I am also going to write this small freeware application, which should appear in 2006, and should later be integrated into iNMR.
- The complete integration into iNMR represents the perfect solution. Whenever you find an unexpected and unknown peak into a spectrum, you'll directly ask the computer: "Where else have you seen such a thing?", and it will open, on the spot, all the old spectra containing a peak at the same position.
You understand that I will be busy in the next few weeks...
Wednesday, 22 November 2006
IUPAC
When we report the chemical shift of a peak, we can use two units, according to IUPAC. The most recent recommendation I have found is:
Pure Appl.Chem., Vol.73, No.11, pp.1795–1818, 2001.
©2001 IUPAC
which defines the δ as:
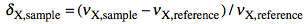
and the Ξ as:

The latter term is the measured 1H frequency of TMS (diluted chloroform solution). The δ scale is limited to a single nuclide, while the Ξ scale is a unified scale valid for all nuclei. The unified scale simplifies the experimental practice for the exotic nuclei, but generates unusual figures with 6 decimal digits. I have always worked with hydrogen and carbon and have never found a chemical shift reported in Ξ units. I am perplexed because IUPAC says:
IUPAC recommends that a unified chemical shift scale for all nuclides be based on the proton resonance of TMS as the primary reference.
and also
In the future, reporting of chemical shift data as Ξ values may become more common and acceptable.
Consider that the above formula for Ξ is only apparently simple: the input values are not normally found inside NMR spectral files. Even if they are, the chemist is not used to calculate the chemical shift: he reads it. To really switch to the unified scale it would be necessary that:
What's really good about the cited article is that it includes all the information on the subject, and we don't have to consult older recommendations (the contrary happened with the definition of JCAMP-DX for NMR, unfortunately). In the following part of this post I will discuss the old δ unit exclusively. Citing again the IUPAC article:
Unfortunately, older software supplied by manufacturers to convert from frequency units to ppm in FT NMR sometimes uses the carrier frequency in the denominator instead of the true frequency of the reference, which can lead to significant errors.
The carrier frequency is the frequency of the transmitter (the center of the spectral width). The reference frequency is the absolute frequency (in the laboratory frame) of the reference compound (TMS, just to materialize the idea). You know that TMS is usually at the far right of the spectrum, so there is a noticeable difference between the two frequencies. When the spectrum leaves the spectrometer, how can you tell which of the two values are exported with the file? I have opened a XWin-NMR file and found:
##$SFO1= 600.1324441176
##$SW= 8.26537126752216
##$SW_h= 4960.31746031746
You can verify that SFO1 = SW_h / SW. _Assuming_ that XWin-NMR follows the IUPAC convention, SFO1 is the frequency of TMS. I don't know what happens with other programs. And I don't know what happens when the user changes the scale reference. According to the rules, if he shifts the scale of even a minimal quantity, let's say 0.001 ppm, the last digits of SFO1 should change.
I know for sure that it is not so with iNMR, which is wrong. iNMR accepts the frequency value found into the original file and keeps it constant (unless the user changes it explicitly). It is possible to recalculate the reference value every time that the TMS position is redefined, but it's almost a paradox. If the users says that the scale is not correct, then other parameters may also be wrong. In order to follow the rule, the program must perform a calculation based on dubious values. If I don't follow the rule, in the common case that the scale is shifted by 0.1 ppm or less, I know to make an error in the order of 10-7. In practice an error of less than 10-4 can be tolerated. Today I prefer to introduce this minimal error, instead of altering the value for the spectrometer frequency. The user is free, however, to set both the value for the spectrometer frequency and the TMS position. Tomorrow, if I change my mind, I can make the process automatic.
Pure Appl.Chem., Vol.73, No.11, pp.1795–1818, 2001.
©2001 IUPAC
which defines the δ as:
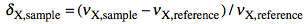
and the Ξ as:

The latter term is the measured 1H frequency of TMS (diluted chloroform solution). The δ scale is limited to a single nuclide, while the Ξ scale is a unified scale valid for all nuclei. The unified scale simplifies the experimental practice for the exotic nuclei, but generates unusual figures with 6 decimal digits. I have always worked with hydrogen and carbon and have never found a chemical shift reported in Ξ units. I am perplexed because IUPAC says:
IUPAC recommends that a unified chemical shift scale for all nuclides be based on the proton resonance of TMS as the primary reference.
and also
In the future, reporting of chemical shift data as Ξ values may become more common and acceptable.
Consider that the above formula for Ξ is only apparently simple: the input values are not normally found inside NMR spectral files. Even if they are, the chemist is not used to calculate the chemical shift: he reads it. To really switch to the unified scale it would be necessary that:
- All journals require the chemical shift expressed in Ξ units.
- All spectrometers have their software updated, so the scale can be optionally be expressed in the new unit (who's going to pay?).
- The absolute frequency of 1H of TMS, measured when installing the instrument, be saved into every file.
- Some standard rule governs the previous point, so all softwares can read spectra from all instruments.
What's really good about the cited article is that it includes all the information on the subject, and we don't have to consult older recommendations (the contrary happened with the definition of JCAMP-DX for NMR, unfortunately). In the following part of this post I will discuss the old δ unit exclusively. Citing again the IUPAC article:
Unfortunately, older software supplied by manufacturers to convert from frequency units to ppm in FT NMR sometimes uses the carrier frequency in the denominator instead of the true frequency of the reference, which can lead to significant errors.
The carrier frequency is the frequency of the transmitter (the center of the spectral width). The reference frequency is the absolute frequency (in the laboratory frame) of the reference compound (TMS, just to materialize the idea). You know that TMS is usually at the far right of the spectrum, so there is a noticeable difference between the two frequencies. When the spectrum leaves the spectrometer, how can you tell which of the two values are exported with the file? I have opened a XWin-NMR file and found:
##$SFO1= 600.1324441176
##$SW= 8.26537126752216
##$SW_h= 4960.31746031746
You can verify that SFO1 = SW_h / SW. _Assuming_ that XWin-NMR follows the IUPAC convention, SFO1 is the frequency of TMS. I don't know what happens with other programs. And I don't know what happens when the user changes the scale reference. According to the rules, if he shifts the scale of even a minimal quantity, let's say 0.001 ppm, the last digits of SFO1 should change.
I know for sure that it is not so with iNMR, which is wrong. iNMR accepts the frequency value found into the original file and keeps it constant (unless the user changes it explicitly). It is possible to recalculate the reference value every time that the TMS position is redefined, but it's almost a paradox. If the users says that the scale is not correct, then other parameters may also be wrong. In order to follow the rule, the program must perform a calculation based on dubious values. If I don't follow the rule, in the common case that the scale is shifted by 0.1 ppm or less, I know to make an error in the order of 10-7. In practice an error of less than 10-4 can be tolerated. Today I prefer to introduce this minimal error, instead of altering the value for the spectrometer frequency. The user is free, however, to set both the value for the spectrometer frequency and the TMS position. Tomorrow, if I change my mind, I can make the process automatic.
Tuesday, 21 November 2006
Mistakes
Apparently a program to process NMR spectra is like any other computer program: eventually they consume ink and paper and, if the user is happy with the printout, he quits the program. The fundamental difference, in the NMR case, is that the user is forced to accept the result even if it slightly wrong. If, for example, there is a very small error in the calculation of integrals, or the spectrum and the scale are misaligned by 1 mm, or the relaxation time has not been correctly estimated, how can the user recognize the mistake? Comparing the results of two different programs is easy, but time-consuming, and not everybody has two programs to compare. This is also what I do as a programmer. Sometimes, when I can't find a second program, I write two different algorithms and test them against each other. Some subtle differences are really difficult to discern; I remember a couple of cases in which it took months. Version 1 of SwaN-MR (the version that nobody used) calculated wrong integrals. The mistake became, however, evident as soon as the program was used in practice.
Version 0.1 of iNMR drew 1D spectra shifted by 1 spectral point (in many cases less than 0.1%). When the scale was calibrated (against the... shifted spectrum), the error was perfectly compensated, because it was constant. There was no way to notice or demonstrate the bug, the spectrum and the scale were perfectly aligned, until I wrote the peak-picking function. The output of peak-picking was constantly shifted by 1 point and in this way the bug was revealed. In the case it was a benign bug with no consequence. My experience is that there is no reason to trust a programmer. The user should find the time to personally test the software, at least those parts that are essential for his work. Remember that they come with no warranty (how could it be different?).
Assuming that the NMR software is perfect, the printed output can still be misleading. Is it possible to tell by inspection if the processing has been performed correctly? The first thing that I observe is the shape of the peaks. It tells if the sample has not been accurately shimmed and if the user relied exclusively on automatic phase correction or, instead, spent the canonical minute in manual phase correction. These things are not as important as baseline correction or accurate referencing against TMS, and have no relation with those other fundamental steps, but are diagnostic hints about the experience and the patience of the user. Including the TMS peak (or a solvent peak) into the peak-picking can be useful to demonstrate that the scale has been correctly referenced. Including the graphical integrals or, better, integrating pieces of pure baseline, can show how flat the latter is. These expedients are not enough, however: small deviations of the integral from zero are not graphically evident and a single baseline sample is only a partial demonstration. The TMS position deserves another article, I hope to write it soon.
Unfortunately the above expedients are not elegant and reduce the clarity and readability of the printed spectrum. What happens, however, if you discover, from the printout, that the processing is inaccurate? When you discover a grammar error in the draft of an article you can correct it and print again. Can you do the same, with the spectrum, in the absence of the raw data? Even if you have an accurate log file of all processing operations, what's it for, if you are not allowed to reprocess the spectrum?
The printed spectrum cannot be replaced because all magnetic and optical supports have been a failure (if their purpose was to save the information for posterity). The CD seems to be more durable than magnetic supports, but it is already being replaced by the DVD and, most of all, we don't have the proof that our CDs will still be readable 20 years from now. Store the spectra on paper, but inspect them on screen, where it is possible to check the quality of processing. The point is that most of casual users lack the basic know-how to judge the quality of a spectrum. The apparent complexity of 2D spectroscopy effectively stops inexperienced users, but in the more familiar 1D environment they feel free to process spectra as they like. Processing 2D spectra is like using a word-processor: the effect of a mistake is apparent even to the uninitiated. The common mistake, in 1D spectroscopy, is to forget that it's a branch of science and that it is to be approached with a minimal knowledge of the field. I admit the sacrosanct right of the user to ignore the manual, but he must already know the steps of routine processing, even when the processing is completely automatic. If he knows how NMR processing works, he can learn the program by trial and error. It's not his/her fault if manuals are boring. My personal advise: read them!
Version 0.1 of iNMR drew 1D spectra shifted by 1 spectral point (in many cases less than 0.1%). When the scale was calibrated (against the... shifted spectrum), the error was perfectly compensated, because it was constant. There was no way to notice or demonstrate the bug, the spectrum and the scale were perfectly aligned, until I wrote the peak-picking function. The output of peak-picking was constantly shifted by 1 point and in this way the bug was revealed. In the case it was a benign bug with no consequence. My experience is that there is no reason to trust a programmer. The user should find the time to personally test the software, at least those parts that are essential for his work. Remember that they come with no warranty (how could it be different?).
Assuming that the NMR software is perfect, the printed output can still be misleading. Is it possible to tell by inspection if the processing has been performed correctly? The first thing that I observe is the shape of the peaks. It tells if the sample has not been accurately shimmed and if the user relied exclusively on automatic phase correction or, instead, spent the canonical minute in manual phase correction. These things are not as important as baseline correction or accurate referencing against TMS, and have no relation with those other fundamental steps, but are diagnostic hints about the experience and the patience of the user. Including the TMS peak (or a solvent peak) into the peak-picking can be useful to demonstrate that the scale has been correctly referenced. Including the graphical integrals or, better, integrating pieces of pure baseline, can show how flat the latter is. These expedients are not enough, however: small deviations of the integral from zero are not graphically evident and a single baseline sample is only a partial demonstration. The TMS position deserves another article, I hope to write it soon.
Unfortunately the above expedients are not elegant and reduce the clarity and readability of the printed spectrum. What happens, however, if you discover, from the printout, that the processing is inaccurate? When you discover a grammar error in the draft of an article you can correct it and print again. Can you do the same, with the spectrum, in the absence of the raw data? Even if you have an accurate log file of all processing operations, what's it for, if you are not allowed to reprocess the spectrum?
The printed spectrum cannot be replaced because all magnetic and optical supports have been a failure (if their purpose was to save the information for posterity). The CD seems to be more durable than magnetic supports, but it is already being replaced by the DVD and, most of all, we don't have the proof that our CDs will still be readable 20 years from now. Store the spectra on paper, but inspect them on screen, where it is possible to check the quality of processing. The point is that most of casual users lack the basic know-how to judge the quality of a spectrum. The apparent complexity of 2D spectroscopy effectively stops inexperienced users, but in the more familiar 1D environment they feel free to process spectra as they like. Processing 2D spectra is like using a word-processor: the effect of a mistake is apparent even to the uninitiated. The common mistake, in 1D spectroscopy, is to forget that it's a branch of science and that it is to be approached with a minimal knowledge of the field. I admit the sacrosanct right of the user to ignore the manual, but he must already know the steps of routine processing, even when the processing is completely automatic. If he knows how NMR processing works, he can learn the program by trial and error. It's not his/her fault if manuals are boring. My personal advise: read them!
Monday, 20 November 2006
Sunday, 19 November 2006
Self-Promotion
Today iNMR is a Universal Binary application.
This is the final stage of a long endeavor. I have added to iNMR all the features asked by hundreds of users of all disciplines and the program is still extremely compact, elegant and immediate. It integrates perfectly with Mac OS X, also because it's the only NMR program which adheres to the Apple guidelines. Now that Apple is selling millions of computers, other software houses are hastily porting their products from Windows, but they can never achieve the same level of integration. Even admitting that these other programs will be ported, they will still look like strangers into Mac OS X.
iNMR is so vast that can be compared to a suite of programs: a processor, an annotator, two modules to simulate both static and dynamic NMR spectra, a versatile line-fitting module, a host of importing filters and another host of exporting functions. To control all these operations you don't have to navigate through long menus or countless palettes, nor to memorize the command names. You have certainly appreciated the clear and simple interface of iNMR.
Despite the wealth of features, most users have never reported a single bug. The few reported errors have been swiftly corrected and a new version has appeared within 24 hours from their discovery. This is certainly the most important thing you wanted to know.
You are kindly invited to download and try the latest version 1.5.6 and to visit the site www.inmr.net for further news.
This is the final stage of a long endeavor. I have added to iNMR all the features asked by hundreds of users of all disciplines and the program is still extremely compact, elegant and immediate. It integrates perfectly with Mac OS X, also because it's the only NMR program which adheres to the Apple guidelines. Now that Apple is selling millions of computers, other software houses are hastily porting their products from Windows, but they can never achieve the same level of integration. Even admitting that these other programs will be ported, they will still look like strangers into Mac OS X.
iNMR is so vast that can be compared to a suite of programs: a processor, an annotator, two modules to simulate both static and dynamic NMR spectra, a versatile line-fitting module, a host of importing filters and another host of exporting functions. To control all these operations you don't have to navigate through long menus or countless palettes, nor to memorize the command names. You have certainly appreciated the clear and simple interface of iNMR.
Despite the wealth of features, most users have never reported a single bug. The few reported errors have been swiftly corrected and a new version has appeared within 24 hours from their discovery. This is certainly the most important thing you wanted to know.
You are kindly invited to download and try the latest version 1.5.6 and to visit the site www.inmr.net for further news.
Practice
Why integral values can never be accurate? I have prepared a set of 2 examples. One contains a single peak in time domain, the other contains a single lorentzian shape. In each case it's possible to alter the linewidth or even to change the shape to gaussian. As usually the required reader is iNMR, which in turn requires Mac OS X. In the time domain example all points are equal to 1. The shape is given exclusively by the weighting function. I have set it to an exponential equivalent to a 10 Hz broadening. After FT the signal is as large as the spectrum, in other words it never reaches zero. There is a significant baseline offset, which actually also contains to the tails of the lorentzian. In the figure below I have already subtracted this offset (the equivalent of multiplying the first point of the FID by 0.5).
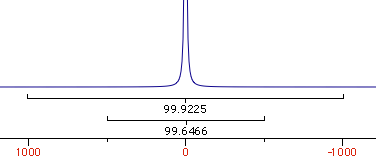
When the integrated region is 100 times the linewidth, the area is still significantly far from the 100%. From the other spectrum I have measured slightly different values, because I have arbitrarily set 100 equal not to the total area, but to the area of a region 2500 times larger than the linewidth.
With your experimental spectra is not the case to define wide integration intervals, like those shown above. Everything works fine because linewidths are comparable and the error is almost constant. Expect to measure lower integrals for wider peaks, even when the relaxation delay is very long.

When the integrated region is 100 times the linewidth, the area is still significantly far from the 100%. From the other spectrum I have measured slightly different values, because I have arbitrarily set 100 equal not to the total area, but to the area of a region 2500 times larger than the linewidth.
| interval/linewidth | area % |
| 10 | 93.6 |
| 20 | 96.8 |
| 100 | 99.4 |
| 250 | 99.7 |
| 500 | 99.9 |
| 5000 | 100 |
With your experimental spectra is not the case to define wide integration intervals, like those shown above. Everything works fine because linewidths are comparable and the error is almost constant. Expect to measure lower integrals for wider peaks, even when the relaxation delay is very long.
Theory
NMR means nuclear magnetic resonance. It' s an analytical technique by which atomic nuclei and, indirectly, their surroundings, are revealed. It's called magnetic because a magnet is used to enhance the (otherwise negligible) energy of the nuclei. It's a resonance phenomenon because the nuclei, when excited by a wave of the right frequency, respond with an analogue wave. The NMR experiment is conceptualized in a rotating coordinate system. For the moment being you just have to pretend it is a plain Cartesian system of coordinates xyz. A radio-frequency pulse tilts the magnetization of the nuclei, initially in the z direction, in the xy plane for detection. Here each nucleus rotates at its own resonance frequency while two detectors sample the total magnetization at regular intervals along the x and y axes. The regular interval is called dwell time. The measured intensities along the x axis are called the real part of the spectrum and intensities along y form the imaginary part. Real and imaginary are just names. They could have been called right and left or red and white and it would have been the same (or better). This is the main difference indeed between an NMR instrument and a hi-fi tuner. It probably serves to justify the price difference. You have to realize that the real and imaginary parts are both true experimental values of the same importance. These intensities are stored on a hard disk in the same time order in which they are sampled (i.e. chronologically). A couple of a real and an imaginary values, collected at the same time, constitute a complex point. iNMR normally displays only the real part of the spectrum. A complex spectrum is like a vector in physics. It can be characterized by its x and y components or by its magnitude (amplitude) and direction. iNMR lets you display the magnitude of the spectrum if you want. The direction of a complex point is called phase. The so called "phase correction" is a process which mixes the real (x) and imaginary (y) components. A radio-frequency wave has frequency, amplitude and phase. Thus complex numbers are the natural choice to describe a RF signal. In an older experimental scheme a single detector measures the magnetization along both axes. In this case the sampling cannot be simultaneous. It is in fact sequential. In this case the spectrum is known as real only. Actually it can (and is) manipulated just as a normal simultaneous, complex, spectrum. A simple ad hoc correction is needed when transforming the spectrum in the "frequency domain".
Let's explain what this last term means. What we have been speaking of up to know is a not very meaningful function of time called FID (free induction decay) (induction is another way to refer to magnetization). It is not very meaningful because all the nuclei resonates at the same time. If there were me and Pavarotti singing together it would be easy for you to discriminate our single contribution to the choir. But you are not trained to discriminate among a collection of atomic nuclei resonating together. The Fourier transformation (FT) is a mathematical tool that separates the contribution of each nucleus by its resonance frequency. The FID is a function of time, while the transformed spectrum is a function of frequency. The FT requires a computer and this is why you need a computer and a software application if you want to do some NMR. At first sight it may seem that the function of frequency should extend between - and +. Actually the sampling theorem states it only has to be calculated in the interval from 0 (included) to Ny (excluded). Ny = Nyquist frequency = sampling rate = reciprocal of the dwell time. Let's take a pause. What's the angle whose sine is 1? My pocket calculator says: 90°. I say: 90°±n360°. Who is right? Both! Coming back to our NMR experiment, suppose a signal is so fast that it rotates by exactly 360° during the dwell time. The two detectors will see it always in the same position, so they will believe it simply doesn't move (it has zero frequency). The same happens with four different signals which rotate by -350°, 10°, 370° and 730° during the dwell time. There is absolutely no way to tell which is which, unless you shorten the dwell time (a common experimental practice). A final case: you have two signals A and B and A moves of 361° respect to B each sampling interval. You will get the impression that A is moving only of 1° each time. In conclusion, the maximum difference in frequency that can be detected is = number of cycles / time interval = 1 / dwell time = Nyquist. q.e.d. In NMR this quantity is called spectral width. All the books report different expressions for the Nyquist frequency and the spectral width. One day we should open a discussion on the subject.
On a purely mathematical basis it doesn't matter how large the actual frequency range you have to record. You can pretend the range starts at zero and extend up to comprise the maximum signal separation. In practice detectors work in the low-frequency range. So you have technical limitations, and this is only the first one. The resonance frequencies today are approaching the GHz. The frame of reference also rotates at a similar frequency, so the apparent frequency is in the range of KHz. With this reduced frequency the dwell time needs to be in the order of milliseconds. The problem is that you need to filter out all other frequencies because they contain nasty noise (didn't I say it was an hi-fi matter?). So we need the rotating frame to move from the ideal world of theory and to become a practical reality. How is the rotating frame accomplished experimentally? A detector receives two signals, one coming from the sample under study and another which is a duplicate of the exciting frequency. The detector actually detects the difference between the two frequencies. To fully exploit the power of the pulse, the transmitter is put at the centre of the spectrum. Signals falling at the left of it appear as negative frequencies. (well, here it is not important if you use the delta or the tau scale and if they are positve or negative; only the concept matters). We have said that the spectrum begins at zero. In fact, if you perform a plain FT, the whole left side would appear shifted by the Nyquist frequency, then to the right of the right side!
The FT and its inverse show a number of interesting properties. The first one predicts that, if you complex-conjugate the FID, the transformed spectrum will be the mirror image of the original spectrum. In fact, if you invert the y component of a vector, you obtain its image across a mirror put along the x axis. Anything rotating counter-clockwise (positive frequency) will appear as rotating clockwise (negative frequency) and vice versa. This mirror image is mathematically called "complex conjugate". Some spectrometers already perform this operation when acquiring. This is another reason (together with sequential acquisition) why spectra coming from different instruments require different processing.
The second property predicts that changing the sign of even points of the FID is equivalent to swapping the left half of the spectrum with the right half. Because they are already swapped and need to be put back in the correct order, you understand this is an useful property. In the beginning of the SwaN-MR era this operation was simply called "swap". A day in which I was sillier than usual I changed the name in "quadrature". A possible explanation is that spectroscopists use to say "I implement quadrature detection" instead of saying "I put the transmitter in the middle of the spectrum". Like conjugation, this operation may have already been performed by the spectrometer during acquisition; in this case don't do it a second time. A third property says that the conjugated FT of a real function is symmetrical. This property is exploited by two techniques known as zero-filling and Hilbert transformation. The exploiting is so indirect and so difficult to explain, that I'll skip the demonstration. Zero-filling means doubling the length of the FID by adding a series of zeroes to its tail. During FT, the information (signal) contained in the imaginary part will fill the empty space. As a final result, you increase the resolution of the spectrum. In fact, the spectral width is already defined by the sampling rate, so adding new points results in increasing their density and the description of details. The Hilbert transformation is the inverse trading: you first zero the imaginary part, then you reconstruct it. Programs present these two operations as different commands and books describe them with different formulae, so most people don't realize that they are two sides of the same coin. There may be cases in which you are forced to zero-fill anyway. It happens that computers prefer to apply the FT only to certain amount of data, precisely to powers of 2. E.g. they like to transform a sequence of 1024 points, but never 1000. In this case 100% of computers will automatically zero-fill to 1024 points without asking your opinion. It's not the case of being fiscal here, I advise you to keep your computer happy.
A fourth property (actually a theorem) says that multiplication in time domain corresponds to convolution in frequency domain. Convolution is crossing two functions. The product equally resembles both parents. Spectroscopists, when they dislike their spectra, use convolutions like breeders cross their cattle. Convolution certainly takes less time than crossing two animals but is still a very long operation in a spectroscopist's perception of time. So it is always preferable to perform a "multiplication in time domain" or, to save three words, "weighting". When you weight you put in practice the Heisenberg uncertainty principle. The longer the apparent life of a signal in time domain, the more resolved it will appear in frequency domain. To reach this goal you multiply the FID with a function which raises in time. At a certain point the true signal is almost completely decayed and the FID only contains noise. When you arrive there it is better to use a function which decays in time in order to reduce the noise. It's the general sort of trading between sensitivity and resolution which makes similar all spectroscopies.
A fifth property says that the total area of the spectrum is equal to the first point in the FID. This property affects both the phase and baseline characteristics of a spectrum. In fact it often happens that phase and baseline distortions are correlated. Now suppose that you have only one signal in your spectrum and that the corresponding magnetization was aligned along the y axis when the first point was sampled. The x (real) part is zero. According to the property, when you FT, half of the signal will be negative for the total area to be zero. Such a spectrum is called a "dispersion" spectrum. Normally an "absorption" spectrum is preferable, because the signal is all positive and narrower and the integral is proportional to the concentration of the chemical species. In the case described the absorption spectrum correspond to the imaginary component. In real-life cases the absorption and dispersion spectra are part in the real and part in the imaginary component. A phase correction separates them and gives the desired absorption spectrum. At last you realize why the NMR signal is recorded in two channels simultaneously! Remember it! The reverse of the coin is that the first point of the spectrum (the one at the transmitter frequency, as explained above) corresponds to the integral of the FID and, because the FID oscillates, to its average position. In theory the latter should be zero so you can decide to raise or lower the FID in order for the average to be exactly zero. In this way you reduce the artifact that the transmitter leaves at the centre of the spectrum. iNMR, by default, does not apply this kind of "DC correction". (DC stays for drift current).
A sixth property says that a time shift corresponds to a linear phase distortion in frequency domain and vice versa. Because you can' t begin acquisition during the exciting pulse (which represents time zero), you normally have to deal with this property. In fact you perform two kinds of correction. The zero-order one, described above, which affects the whole spectrum uniformly, and a first-order one, whose effect varies linearly with frequency. If in turn you want to shift all your frequencies by a non-integer number of points, the best way is to apply a first-order phase change to the FID.
Finally the Fourier transformation shares the general properties of linear functions: it commutes with multiplication by a constant and with addition. Sometimes FT alone is not enough to separate signals. In this cases the acquisition scheme is complicated with the addition of delays and pulses. During a delay, for example, two signal starting with the same phase but rotating at different frequencies lose their phase equality. A pulse has no effect on a signal aligned along its axis but rotates a signal perpendicular (in phase quadrature) to it. So you should not wonder that a suitable combination of pulses and delays can differentiate between signals. A signal is in effect a quantic transition between two energy levels A and B, caused by a RF pulse. A second pulse can move the transition to a third level C. It will be a new transition with a new frequency. This is just to show how many things can be done. In the simplest experiment the FID is a simple function of time f(t). If we introduce a delay d at any point before the acquisition the FID becomes f(d,t). Now if we run many times the experiment, each time with a different value of d, d becomes a new time variable indeed. So the expression for the FID is now f(t1, t2). t2 comes later in the experimental scheme, so it corresponds to the "t" of the simple 1-pulse experiment. What do you have on the hard disk? A rectangular, ordered set of complex points called matrix. The methods used to display it are usually borrowed by geography. If two dimensions are not enough for you, you can add a third or even a fourth one. The dimension with the highest index is said to be directly revealed because switching on the detectors is the last stage in a pulse sequence.
To go from FID(t1,t2) to Spectrum(f1, f2) you need to do everything twice. First weight, zero-fill, FT and phase correct along t2, then weight, zero-fill and phase correct along f1. Finally you can correct the baseline. The only freedom iNMR gives to you outside this scheme is that you can also correct the phase along f2 after the correction along f1. This partial lack of freedom simplifies immensely your work. Now there are good news and bad news. Bad news first. In 1-D spectroscopy you appreciated the need of acquiring the spectrum along two channels in quadrature. The books says it is required in order to put the transmitter in the middle of the spectrum. You know it is false. The true reason is that quadrature detection is needed to see the spectrum in absorption mode. The same holds for the f1 dimension. You need to duplicate everything again. Instead of using an hypercomplex point (which would need a 4D coordinate system to be conceptualised) it is customary to store points in their chronological order. You also have the real component of f1 stored in the odd rows of the matrix and the imaginary component in the even rows. The idea is that, after the spectrum is transformed and in absorption along f2, you can discard the imaginary part, because it only contains dispersion, and merge each odd row with the subsequent. After the merging your points are complex again (and the number of rows is halved). The real part is the same as before while what was the real part of even rows is now the imaginary part. After the final FT the spectrum will be phaseable along f1 (but not along f2). In case you want to perform all phase corrections at the end, do not throw away the imaginary part and process it separately. When you need to correct the phase along f2, you swap the imaginary part along f1 with the imaginary part along f2. Now the good news: iNMR does all the book-keeping for you! You just have to specify if you want to proceed with the minimal amount of data or if you prefer keeping all of it. The scheme outlined above is called Ruben-States. Another scheme exists, called TPPI. Everything was said holds, plus TPPI requires a real FT, plus it already negates even points (you have to remember of not doing it again). Just because two is not the perfect number, someone added a third scheme called States-TPPI. Fortunately it is the simplest solution, in that it requires a plain FT (no "quadrature" like Ruben-States). The most ancient (and probably the most useful) 2D experiment, the COSY experiment, is not phaseable because it is even older than the oldest scheme. So you don't need to bother with all these things. Just switch to magnitude representation. To be honest, processing a COSY experiment is certainly easier than processing a 1D proton spectrum. There is also no need to correct the baseline and less sense in measuring integrals. Another widely used protocol is echo-antiecho. It's slightly more complicated and had the merit of allowing new phase-sensitive experiments, starting with HSQC (hetero-nuclear correlation detected through the hydrogen magnetization).
If this long speech was too complicated for your tastes, it doesn't mean you cannot become an expert in NMR processing. Simply playing with iNMR, by trial and error, observing the effect of each option and pouring a small dose of common sense you can still become a wizard.
Let's explain what this last term means. What we have been speaking of up to know is a not very meaningful function of time called FID (free induction decay) (induction is another way to refer to magnetization). It is not very meaningful because all the nuclei resonates at the same time. If there were me and Pavarotti singing together it would be easy for you to discriminate our single contribution to the choir. But you are not trained to discriminate among a collection of atomic nuclei resonating together. The Fourier transformation (FT) is a mathematical tool that separates the contribution of each nucleus by its resonance frequency. The FID is a function of time, while the transformed spectrum is a function of frequency. The FT requires a computer and this is why you need a computer and a software application if you want to do some NMR. At first sight it may seem that the function of frequency should extend between - and +. Actually the sampling theorem states it only has to be calculated in the interval from 0 (included) to Ny (excluded). Ny = Nyquist frequency = sampling rate = reciprocal of the dwell time. Let's take a pause. What's the angle whose sine is 1? My pocket calculator says: 90°. I say: 90°±n360°. Who is right? Both! Coming back to our NMR experiment, suppose a signal is so fast that it rotates by exactly 360° during the dwell time. The two detectors will see it always in the same position, so they will believe it simply doesn't move (it has zero frequency). The same happens with four different signals which rotate by -350°, 10°, 370° and 730° during the dwell time. There is absolutely no way to tell which is which, unless you shorten the dwell time (a common experimental practice). A final case: you have two signals A and B and A moves of 361° respect to B each sampling interval. You will get the impression that A is moving only of 1° each time. In conclusion, the maximum difference in frequency that can be detected is = number of cycles / time interval = 1 / dwell time = Nyquist. q.e.d. In NMR this quantity is called spectral width. All the books report different expressions for the Nyquist frequency and the spectral width. One day we should open a discussion on the subject.
On a purely mathematical basis it doesn't matter how large the actual frequency range you have to record. You can pretend the range starts at zero and extend up to comprise the maximum signal separation. In practice detectors work in the low-frequency range. So you have technical limitations, and this is only the first one. The resonance frequencies today are approaching the GHz. The frame of reference also rotates at a similar frequency, so the apparent frequency is in the range of KHz. With this reduced frequency the dwell time needs to be in the order of milliseconds. The problem is that you need to filter out all other frequencies because they contain nasty noise (didn't I say it was an hi-fi matter?). So we need the rotating frame to move from the ideal world of theory and to become a practical reality. How is the rotating frame accomplished experimentally? A detector receives two signals, one coming from the sample under study and another which is a duplicate of the exciting frequency. The detector actually detects the difference between the two frequencies. To fully exploit the power of the pulse, the transmitter is put at the centre of the spectrum. Signals falling at the left of it appear as negative frequencies. (well, here it is not important if you use the delta or the tau scale and if they are positve or negative; only the concept matters). We have said that the spectrum begins at zero. In fact, if you perform a plain FT, the whole left side would appear shifted by the Nyquist frequency, then to the right of the right side!
The FT and its inverse show a number of interesting properties. The first one predicts that, if you complex-conjugate the FID, the transformed spectrum will be the mirror image of the original spectrum. In fact, if you invert the y component of a vector, you obtain its image across a mirror put along the x axis. Anything rotating counter-clockwise (positive frequency) will appear as rotating clockwise (negative frequency) and vice versa. This mirror image is mathematically called "complex conjugate". Some spectrometers already perform this operation when acquiring. This is another reason (together with sequential acquisition) why spectra coming from different instruments require different processing.
The second property predicts that changing the sign of even points of the FID is equivalent to swapping the left half of the spectrum with the right half. Because they are already swapped and need to be put back in the correct order, you understand this is an useful property. In the beginning of the SwaN-MR era this operation was simply called "swap". A day in which I was sillier than usual I changed the name in "quadrature". A possible explanation is that spectroscopists use to say "I implement quadrature detection" instead of saying "I put the transmitter in the middle of the spectrum". Like conjugation, this operation may have already been performed by the spectrometer during acquisition; in this case don't do it a second time. A third property says that the conjugated FT of a real function is symmetrical. This property is exploited by two techniques known as zero-filling and Hilbert transformation. The exploiting is so indirect and so difficult to explain, that I'll skip the demonstration. Zero-filling means doubling the length of the FID by adding a series of zeroes to its tail. During FT, the information (signal) contained in the imaginary part will fill the empty space. As a final result, you increase the resolution of the spectrum. In fact, the spectral width is already defined by the sampling rate, so adding new points results in increasing their density and the description of details. The Hilbert transformation is the inverse trading: you first zero the imaginary part, then you reconstruct it. Programs present these two operations as different commands and books describe them with different formulae, so most people don't realize that they are two sides of the same coin. There may be cases in which you are forced to zero-fill anyway. It happens that computers prefer to apply the FT only to certain amount of data, precisely to powers of 2. E.g. they like to transform a sequence of 1024 points, but never 1000. In this case 100% of computers will automatically zero-fill to 1024 points without asking your opinion. It's not the case of being fiscal here, I advise you to keep your computer happy.
A fourth property (actually a theorem) says that multiplication in time domain corresponds to convolution in frequency domain. Convolution is crossing two functions. The product equally resembles both parents. Spectroscopists, when they dislike their spectra, use convolutions like breeders cross their cattle. Convolution certainly takes less time than crossing two animals but is still a very long operation in a spectroscopist's perception of time. So it is always preferable to perform a "multiplication in time domain" or, to save three words, "weighting". When you weight you put in practice the Heisenberg uncertainty principle. The longer the apparent life of a signal in time domain, the more resolved it will appear in frequency domain. To reach this goal you multiply the FID with a function which raises in time. At a certain point the true signal is almost completely decayed and the FID only contains noise. When you arrive there it is better to use a function which decays in time in order to reduce the noise. It's the general sort of trading between sensitivity and resolution which makes similar all spectroscopies.
A fifth property says that the total area of the spectrum is equal to the first point in the FID. This property affects both the phase and baseline characteristics of a spectrum. In fact it often happens that phase and baseline distortions are correlated. Now suppose that you have only one signal in your spectrum and that the corresponding magnetization was aligned along the y axis when the first point was sampled. The x (real) part is zero. According to the property, when you FT, half of the signal will be negative for the total area to be zero. Such a spectrum is called a "dispersion" spectrum. Normally an "absorption" spectrum is preferable, because the signal is all positive and narrower and the integral is proportional to the concentration of the chemical species. In the case described the absorption spectrum correspond to the imaginary component. In real-life cases the absorption and dispersion spectra are part in the real and part in the imaginary component. A phase correction separates them and gives the desired absorption spectrum. At last you realize why the NMR signal is recorded in two channels simultaneously! Remember it! The reverse of the coin is that the first point of the spectrum (the one at the transmitter frequency, as explained above) corresponds to the integral of the FID and, because the FID oscillates, to its average position. In theory the latter should be zero so you can decide to raise or lower the FID in order for the average to be exactly zero. In this way you reduce the artifact that the transmitter leaves at the centre of the spectrum. iNMR, by default, does not apply this kind of "DC correction". (DC stays for drift current).
A sixth property says that a time shift corresponds to a linear phase distortion in frequency domain and vice versa. Because you can' t begin acquisition during the exciting pulse (which represents time zero), you normally have to deal with this property. In fact you perform two kinds of correction. The zero-order one, described above, which affects the whole spectrum uniformly, and a first-order one, whose effect varies linearly with frequency. If in turn you want to shift all your frequencies by a non-integer number of points, the best way is to apply a first-order phase change to the FID.
Finally the Fourier transformation shares the general properties of linear functions: it commutes with multiplication by a constant and with addition. Sometimes FT alone is not enough to separate signals. In this cases the acquisition scheme is complicated with the addition of delays and pulses. During a delay, for example, two signal starting with the same phase but rotating at different frequencies lose their phase equality. A pulse has no effect on a signal aligned along its axis but rotates a signal perpendicular (in phase quadrature) to it. So you should not wonder that a suitable combination of pulses and delays can differentiate between signals. A signal is in effect a quantic transition between two energy levels A and B, caused by a RF pulse. A second pulse can move the transition to a third level C. It will be a new transition with a new frequency. This is just to show how many things can be done. In the simplest experiment the FID is a simple function of time f(t). If we introduce a delay d at any point before the acquisition the FID becomes f(d,t). Now if we run many times the experiment, each time with a different value of d, d becomes a new time variable indeed. So the expression for the FID is now f(t1, t2). t2 comes later in the experimental scheme, so it corresponds to the "t" of the simple 1-pulse experiment. What do you have on the hard disk? A rectangular, ordered set of complex points called matrix. The methods used to display it are usually borrowed by geography. If two dimensions are not enough for you, you can add a third or even a fourth one. The dimension with the highest index is said to be directly revealed because switching on the detectors is the last stage in a pulse sequence.
To go from FID(t1,t2) to Spectrum(f1, f2) you need to do everything twice. First weight, zero-fill, FT and phase correct along t2, then weight, zero-fill and phase correct along f1. Finally you can correct the baseline. The only freedom iNMR gives to you outside this scheme is that you can also correct the phase along f2 after the correction along f1. This partial lack of freedom simplifies immensely your work. Now there are good news and bad news. Bad news first. In 1-D spectroscopy you appreciated the need of acquiring the spectrum along two channels in quadrature. The books says it is required in order to put the transmitter in the middle of the spectrum. You know it is false. The true reason is that quadrature detection is needed to see the spectrum in absorption mode. The same holds for the f1 dimension. You need to duplicate everything again. Instead of using an hypercomplex point (which would need a 4D coordinate system to be conceptualised) it is customary to store points in their chronological order. You also have the real component of f1 stored in the odd rows of the matrix and the imaginary component in the even rows. The idea is that, after the spectrum is transformed and in absorption along f2, you can discard the imaginary part, because it only contains dispersion, and merge each odd row with the subsequent. After the merging your points are complex again (and the number of rows is halved). The real part is the same as before while what was the real part of even rows is now the imaginary part. After the final FT the spectrum will be phaseable along f1 (but not along f2). In case you want to perform all phase corrections at the end, do not throw away the imaginary part and process it separately. When you need to correct the phase along f2, you swap the imaginary part along f1 with the imaginary part along f2. Now the good news: iNMR does all the book-keeping for you! You just have to specify if you want to proceed with the minimal amount of data or if you prefer keeping all of it. The scheme outlined above is called Ruben-States. Another scheme exists, called TPPI. Everything was said holds, plus TPPI requires a real FT, plus it already negates even points (you have to remember of not doing it again). Just because two is not the perfect number, someone added a third scheme called States-TPPI. Fortunately it is the simplest solution, in that it requires a plain FT (no "quadrature" like Ruben-States). The most ancient (and probably the most useful) 2D experiment, the COSY experiment, is not phaseable because it is even older than the oldest scheme. So you don't need to bother with all these things. Just switch to magnitude representation. To be honest, processing a COSY experiment is certainly easier than processing a 1D proton spectrum. There is also no need to correct the baseline and less sense in measuring integrals. Another widely used protocol is echo-antiecho. It's slightly more complicated and had the merit of allowing new phase-sensitive experiments, starting with HSQC (hetero-nuclear correlation detected through the hydrogen magnetization).
If this long speech was too complicated for your tastes, it doesn't mean you cannot become an expert in NMR processing. Simply playing with iNMR, by trial and error, observing the effect of each option and pouring a small dose of common sense you can still become a wizard.
Saturday, 18 November 2006
Interface
It would be fantastic if we could use instinctively all the software we find on our computers. It's a necessity too: new programs appear every week. The solution is to stop thinking at the programs as products or tools, and even ignore their existence. This is an old, current and welcome trend: the tool is the computer. It comes with its own commands (Open, Quit, Copy, Paste, Print...) and its own graphic style. When you have learned to use your first program, there is nothing more to learn. You will always find the same commands at the same places. After a few days you don't even have to think, because your fingers remember what to do for you. The impossible part is to stick to the same operative system for a lifetime, like we can do with a chronograph, a pen or a turntable (if we are affectioned to them). The difficult part is to switch to a different OS. On Windows you find many tiny icons, on the Mac a few large icons, on Linux all kinds of distros. The (unfortunate) unifying trend is to make longer and longer menus. Menu commands are considered a remnant of the past, like the command line, and left to expert users. What's the fault of expert users, that are faced with these endless menus? It is simpler to type the command on the command line than to find it into a list of 100 items. Even icons are a tragedy. To increase the intensity of the peaks some genius had the idea of creating a "plus" icon that the user was expected to click with the mouse. Afterwards they introduced specific keys, on the keyboard, to control the volume of loudspeakers. You can't find, unfortunately, NMR-specific keyboards. If your NMR program doesn't implement any command line, the keyboard is apparently useless. No law prevents, however, the software to accept input through intuitive keys, like + - or the arrow keys. With a small dose of inventiveness z can mean "zoom", i can mean "integrate", g means "show and hide the grid", p and H set the scale units to ppm or Hz, etc... In other fields (games, music notation) such reconversion of the keyboard is normal. You see that, eventually, all programs are forced to invent something new, and their users are forced to study the manual. What happens when the same program runs on many operative systems? Should it always have the same interface or adapt itself as a chameleon? The first option is the favorite one by Java programmers and saves a lot of efforts, first of all the effort or reading the interface guidelines valid for each system. Apparently it comes at the user's advantage: she can switch to any computer, and still find the same NMR interface. Just a little doubt: if she is forced to switch to another computer, how many chances are she will find the same, familiar, NMR software?
In practice elegance and enjoyment are the decisive factors, especially in the lucky case that the users can choose the software to buy. Windows users want a program with a Windows look-and-feel, and let's not speak of Mac users. They are fanatic purists. An atypical and forgotten case was the first program written by Tecmag, in the early 90s. It was called MacFID and intentionally tried to recreate the complexity of an old-time spectrometer, with dozens of virtual knobs, and list of parameters instead of dialogs. It's not a case that it soon disappeared. If somebody buys a Mac, it' s because she wants an elegant computer with elegant software inside.
Most of the NMR programs allow the personalization of the interface. It's funny to make a tour of a department and observe all the variations that have been created. They are the main reason why the command line is still so widely popular among technician. When they give assistance, how could they find the menu commands or the icons if every spectrometer shows a different graphic interface?
In practice elegance and enjoyment are the decisive factors, especially in the lucky case that the users can choose the software to buy. Windows users want a program with a Windows look-and-feel, and let's not speak of Mac users. They are fanatic purists. An atypical and forgotten case was the first program written by Tecmag, in the early 90s. It was called MacFID and intentionally tried to recreate the complexity of an old-time spectrometer, with dozens of virtual knobs, and list of parameters instead of dialogs. It's not a case that it soon disappeared. If somebody buys a Mac, it' s because she wants an elegant computer with elegant software inside.
Most of the NMR programs allow the personalization of the interface. It's funny to make a tour of a department and observe all the variations that have been created. They are the main reason why the command line is still so widely popular among technician. When they give assistance, how could they find the menu commands or the icons if every spectrometer shows a different graphic interface?
Can't be pregnancy
A reader complains because MestreNova hasn't appeared yet, despite the web site says: "alpha version planned for 15th November". As you may know, a program can have countless alpha and beta versions, and they certainly have some kind of alpha version already compiled. We don't know however, what they mean with "alpha", and if they also plan to make it public. I suppose that the reader who wrote: "we are waiting for mestrenova... the date of release was 15/11 but it isn't ready!!!" is also an old-time customer of Mestre-C, because the first phases of testing will likely be restricted to old Mestre-C users. This is what my friends at Mestrelab confirmed to me in September. One may observe that, normally, alpha versions remain internal to the software house, while beta versions (either public or not) are submitted to the end-users. The web announcement is misleading, because lets you believe that the alpha version will be publicly available (admirable). Certainly it is outdated, by now. They have chosen the strategy of a premature announcement and now have added more tension: all they wanted was to be noticed and they hit the target. This is certainly _not_ the kind of delay you should worry about.
iNMR UB is receiving less attention, because it's simply a recompilation. The program was fast enough even when ran on Rosetta, so fast that they bought it nonetheless. The testing won't be long. All the reports, up to now, say that this new version is just perfect. If nothing new happens, version 1.5.6 will officially debut on monday. What's really new about it is the lower price.
iNMR UB is receiving less attention, because it's simply a recompilation. The program was fast enough even when ran on Rosetta, so fast that they bought it nonetheless. The testing won't be long. All the reports, up to now, say that this new version is just perfect. If nothing new happens, version 1.5.6 will officially debut on monday. What's really new about it is the lower price.
Friday, 17 November 2006
Phase Tutorial
If you want to repeat this tutorial, you need Mac OS 10.3, iNMR and this large 2D spectrum (5.7 MB). You don't need to buy a license (it is only required for printing). If you don't have a Mac, look at the pictures below. After installing iNMR and decompressing the gzip archive, from inside the program open the spectrum. You'll see it in time domain. The command "Tools/Dashboard" opens a palette with the frequently used tools. Only a few are initially enabled. This one:

starts the engine. You need to click it twice, because it's a 2D spectrum. The engine performs weighting and the Fourier Transform. Optionally it can also perform solvent suppression and linear prediction. After the two clicks, you have the transformed spectrum, with no phase correction. All peaks look like:
Why the engine forgets to correct the phase? The intention of the engine was not to automatize all tasks. Quite simply, many operations were (almost) always performed with default parameters. It was a non-sense to perform manually exactly the same operations with the same parameters, therefore the "gears" button was introduced. Phase correction is less repetitive, instead. This is a TOCSY spectrum acquired on a Bruker Avance. The needed phase correction, in this particular case, is well known. Click the icon for manual phase correction:

When the dialog appears, enter the values below:
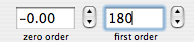
These values are the ones most frequently used, for many kinds of experiments. Another frequent combination is: 9O; 180. We can say the phase is OK along the horizontal dimension (f1, in this case), because all the peaks appear as thin vertical stripes.
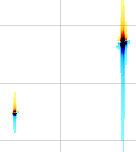
The purpose of phase correction is to have narrow peaks. In order to have our peaks narrow along both directions, we select the "vertical" radio button, as in the picture:
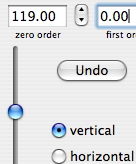
Move the left slider until the zero order correction is = 119 degrees. You should notice an improvement in the center of the spectrum. We don't want to lose this local improvement. Pressing the "alt" key, click near 5 ppm. This mark appears:

It fixes the pivot point. Now the first order phase correction cannot change the row at 5.142 ppm. Press the key "+" to enhance all peaks. Those in the upper-right corner are still not symmetric because they have both positive (red) and negative (blue) tails.
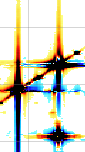
We only need a small adjustment. Move the central slider to "Fine".
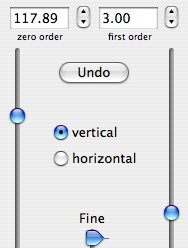
Monitor the situation along the diagonal. The central peak in the figure below is optimal for monitoring, because there is no cross-peak near it. When both tails have the same color, the phase is perfect.
Such easy examples are quite common. They are ideal for training. Try altering the phase and correcting it again. The experience you gain in this way will be precious for tackling more difficult cases.

starts the engine. You need to click it twice, because it's a 2D spectrum. The engine performs weighting and the Fourier Transform. Optionally it can also perform solvent suppression and linear prediction. After the two clicks, you have the transformed spectrum, with no phase correction. All peaks look like:

Why the engine forgets to correct the phase? The intention of the engine was not to automatize all tasks. Quite simply, many operations were (almost) always performed with default parameters. It was a non-sense to perform manually exactly the same operations with the same parameters, therefore the "gears" button was introduced. Phase correction is less repetitive, instead. This is a TOCSY spectrum acquired on a Bruker Avance. The needed phase correction, in this particular case, is well known. Click the icon for manual phase correction:

When the dialog appears, enter the values below:

These values are the ones most frequently used, for many kinds of experiments. Another frequent combination is: 9O; 180. We can say the phase is OK along the horizontal dimension (f1, in this case), because all the peaks appear as thin vertical stripes.

The purpose of phase correction is to have narrow peaks. In order to have our peaks narrow along both directions, we select the "vertical" radio button, as in the picture:

Move the left slider until the zero order correction is = 119 degrees. You should notice an improvement in the center of the spectrum. We don't want to lose this local improvement. Pressing the "alt" key, click near 5 ppm. This mark appears:

It fixes the pivot point. Now the first order phase correction cannot change the row at 5.142 ppm. Press the key "+" to enhance all peaks. Those in the upper-right corner are still not symmetric because they have both positive (red) and negative (blue) tails.

We only need a small adjustment. Move the central slider to "Fine".

Monitor the situation along the diagonal. The central peak in the figure below is optimal for monitoring, because there is no cross-peak near it. When both tails have the same color, the phase is perfect.

Such easy examples are quite common. They are ideal for training. Try altering the phase and correcting it again. The experience you gain in this way will be precious for tackling more difficult cases.
Wednesday, 15 November 2006
Never in Time
I started writing iNMR when 90% of Mac users had already switched to OS X, after Varian had already introduced VNMRJ for Mac OS X. I felt they had invaded my territory and, partly prompted from this challenge, decided to compete.
After one month Steve Jobs announced the next migration, from PowerPC to Intel. Then I knew I was working for a short-lived computer, the PPC iMac. A few months later, another program appeared for OS X, called NMR notebook. In January 2007 the MacBook arrived on the shelves. When iNMR 1.0 shipped, it was already old, because it was not Universal Binary. At the same time the public complained because it had less functions than SwaN-MR. And there were already two competiors. If I had sold as little as 200 copies I had certainly recompiled iNMR on the spot. Adding the desired functions required more effort but less money, and this is what I have done during 2006. Yesterday I began writing the UB version and today it was ready. It's OK, but it takes too long for a deep testing. It's unbelievable the wealth of features contained into a floppy (this is the size of the download). This evening I published iNMR UB in the form of a beta version, hoping to find some collaboration for testing. The beta version requires system 10.4, but the final version will be compatible with 10.3. What else can happen before it becomes an official release? Acorn and Mestrelab have already announced their respective versions for OS X. It's impossible to find a weak spot into the products of the competitors, if they keep these products well closed into their drawers. The winner is always Apple, which managed to sell two computers per programmer: the UB version must necessarily be tested on two different computers.
After one month Steve Jobs announced the next migration, from PowerPC to Intel. Then I knew I was working for a short-lived computer, the PPC iMac. A few months later, another program appeared for OS X, called NMR notebook. In January 2007 the MacBook arrived on the shelves. When iNMR 1.0 shipped, it was already old, because it was not Universal Binary. At the same time the public complained because it had less functions than SwaN-MR. And there were already two competiors. If I had sold as little as 200 copies I had certainly recompiled iNMR on the spot. Adding the desired functions required more effort but less money, and this is what I have done during 2006. Yesterday I began writing the UB version and today it was ready. It's OK, but it takes too long for a deep testing. It's unbelievable the wealth of features contained into a floppy (this is the size of the download). This evening I published iNMR UB in the form of a beta version, hoping to find some collaboration for testing. The beta version requires system 10.4, but the final version will be compatible with 10.3. What else can happen before it becomes an official release? Acorn and Mestrelab have already announced their respective versions for OS X. It's impossible to find a weak spot into the products of the competitors, if they keep these products well closed into their drawers. The winner is always Apple, which managed to sell two computers per programmer: the UB version must necessarily be tested on two different computers.
Monday, 13 November 2006
Vigil
Mestrelab has announced that, during this week, they will reveal to the world the alpha version of MestreNova. Nobody has seen it yet. Everybody seems to know Mestre-C, instead, and I will analyze it. MestreNova has been written from scratch: it can be considered a brand new product whose function is to supersede Mestre-C. The makers had concluded that the old program needed drastic changes, and went for what I always consider the best solution: a fresh new start. Varian preferred to put a new dress over VNMR, and created VNMRJ, which is a dress more than a computer program. Even TopSpin probably contains parts of older programs (some part probably even predates XWin-NMR). Mestrelab has assured me that their new program is 100% new. I hope not to found the bugs and, most of all, the heavy mentality of Mestre-C.
A not so little detail gives the idea of what Mestre-C has always been, namely an unfinished product. Let's say you have assigned and annotated your spectrum: near each peak there is a label pointing to the corresponding atom. You worked for 30 minutes, the results looks nice. There is a multiplet you want to observe in detail. You start zooming in and panning. The spectrum becomes a mess. When you realize that all annotations have changed position, it's too late for undoing. The undo command doesn't work. There is a bug, never corrected, and the effect is that the annotations don't remember their position. You say: "OK, I'll think about it later; now I hide the annotations". Sorry, there is no command to hide the annotations. The best thing you can do is to close the document without saving the changes and open it again. This has been my experience and the experience of the colleagues I have talked with. How do you call it: user-unfriendliness? Using Mestre-C is like dealing with a known thief; the difference is that, instead of keeping your hands into your pockets, you save very frequently, to prevent damages like this. Certainly there is the Undo command, but it is not constantly enabled. It may happen that you can undo a processing command (I prefer reloading the FID, and I have plenty of serious reasons for preferring it) but if you press the wrong key or click the wrong icon, you can't undo.
Mestre-C is very popular because it was the first free NMR software for Windows, has been adequately promoted, and the migration to a commercial product has been gradual. Until last year you could still download a new version every time your old demo had expired. What fascinated the organic chemists is that the authors were other organic chemists. The absence of a price, and the initial lack of experience, had, unfortunately, their natural consequences. The growth of Mestre-C has been slow (taking nearly a decade) and tormented. The chemists could not switch to it, because it was not complete, and considered Mestre-C like a useful tool for special purposes. Eventually they were not interested into switching anymore (I mean: adopting Mestre-C as their exclusive NMR software). They cared very little of the basic functionalities. They already had VNMR or XWin-NMR for everyday tasks. They used Mestre-C as an extension. The number of users reached 20000 long ago, but the number of switchers is very limited, in comparison. The few switchers that I know (myself included) are unhappy. It can be our fault, or a fault of our PCs (all of them are black Dells). Let me bring this example: another absurd bug creates curious and inexplicable ghost splittings. A singlet can show, on the screen only, a perceptible splitting that can be interpreted as a small coupling. When printed, however, it is clearly a singlet. Does it depend on the graphic card, on the operative system or on the program?
The heavy approach of Mestre-C is not a bug, it's a deliberate choice. I can't say that it doesn't work, but I don't like it at all. I prefer doing things with a soft approach and I _feel_ that the heavy approach is _completely_ wrong. Heavy means that:
The word "defaults" recalls another bug that hits me: every time I update the program I lose my personal preferences. Unfortunately I never remember how to set them. There are too many options, most of them undesired and not understood, they are located in many places, some of them are document-specific, others are application-global. A possible cause of my disorientation can be the fact that I don't like Windows. In my former group, most of the members, like me, came from the Macintosh and SwaN-MR. The year before our company forced us to switch to Windows, it had happened that a young researcher had joined our group and she found similar difficulties, because she was switching from Windows/Mestre-C to Mac/SwaN-MR. There were two fundamental differences, though:
Can you see why I am curious to see MestreNova? Don't believe that the rest of Mestre-C was OK. Remember that there is a space for comments here below, so you can correct me if I am wrong, or add your personal experience. Regarding mine, it was limited to what I call "routine processing". Nor is it simple to learn the rest. Every command adds its own peculiar interface. You have to remember what a right-clicks performs under each circumstance. Even when the interface is the same, a surprise can arrive like a punch in the stomach. Phase correction, for example, has a unified interface in 1D and 2D. In the latter case I can't specify a pivot point. Why? I ignore it. I only know that it's hard to correct the phase without a pivot. To continue the torture, in 2D, you also have plenty of confusing colors.
Of course I have always expressed, along the years, my criticism to the author, detailing it more than today. The usual answer was the unarguable: "Customers like it in this way". Another common, and not acceptable, answer was: "The bug will disappear with MestreNova". The most important answer, although a silent one, is MestreNova itself: the fact that they have rewritten it all sounds like a condemnation to death for Mestre-C. Let us remember the good qualities of the defunct. The manual, a little outdated, is readable and helpful. The printed output is OK. The job, eventually, is done. Without enjoyment, yet done. For some users it can also be great to know that there are so many advanced options in store.
I am curious: will everything change with the advent of MestreNova? If the answer is yes, will the users be happy to change their habits, to learn new commands, to adopt a new mentality? Most of all: will the heavy approach survive?
A not so little detail gives the idea of what Mestre-C has always been, namely an unfinished product. Let's say you have assigned and annotated your spectrum: near each peak there is a label pointing to the corresponding atom. You worked for 30 minutes, the results looks nice. There is a multiplet you want to observe in detail. You start zooming in and panning. The spectrum becomes a mess. When you realize that all annotations have changed position, it's too late for undoing. The undo command doesn't work. There is a bug, never corrected, and the effect is that the annotations don't remember their position. You say: "OK, I'll think about it later; now I hide the annotations". Sorry, there is no command to hide the annotations. The best thing you can do is to close the document without saving the changes and open it again. This has been my experience and the experience of the colleagues I have talked with. How do you call it: user-unfriendliness? Using Mestre-C is like dealing with a known thief; the difference is that, instead of keeping your hands into your pockets, you save very frequently, to prevent damages like this. Certainly there is the Undo command, but it is not constantly enabled. It may happen that you can undo a processing command (I prefer reloading the FID, and I have plenty of serious reasons for preferring it) but if you press the wrong key or click the wrong icon, you can't undo.
Mestre-C is very popular because it was the first free NMR software for Windows, has been adequately promoted, and the migration to a commercial product has been gradual. Until last year you could still download a new version every time your old demo had expired. What fascinated the organic chemists is that the authors were other organic chemists. The absence of a price, and the initial lack of experience, had, unfortunately, their natural consequences. The growth of Mestre-C has been slow (taking nearly a decade) and tormented. The chemists could not switch to it, because it was not complete, and considered Mestre-C like a useful tool for special purposes. Eventually they were not interested into switching anymore (I mean: adopting Mestre-C as their exclusive NMR software). They cared very little of the basic functionalities. They already had VNMR or XWin-NMR for everyday tasks. They used Mestre-C as an extension. The number of users reached 20000 long ago, but the number of switchers is very limited, in comparison. The few switchers that I know (myself included) are unhappy. It can be our fault, or a fault of our PCs (all of them are black Dells). Let me bring this example: another absurd bug creates curious and inexplicable ghost splittings. A singlet can show, on the screen only, a perceptible splitting that can be interpreted as a small coupling. When printed, however, it is clearly a singlet. Does it depend on the graphic card, on the operative system or on the program?
The heavy approach of Mestre-C is not a bug, it's a deliberate choice. I can't say that it doesn't work, but I don't like it at all. I prefer doing things with a soft approach and I _feel_ that the heavy approach is _completely_ wrong. Heavy means that:
- The processed spectrum overwrites the FID and the latter is lost.
- Processing is not alterable. If you want to change a window function, for example, you have to restart from the raw data: you lose all of your processing.
- Instead of creating (light) links among spectra, you have to put all the spectra into a single file.
- To create an inset, you are forced to duplicate the spectrum. Each inset, no matter how small, eats as much computer memory and disc space as the whole spectrum.
- The document does not correspond to a spectrum. It is instead a container, which can be empty or contain a number of objects. An object can be a spectrum or any other thing. Even if you avoid such complications and always keep a single spectrum/object into each window, you see a frame around the spectrum, with four handles. I myself keep clicking and dragging the frame by mistake and find it extremely annoying.
- When I add 1D projections along the sides of a 2D plot, I see additional frames.
- By default I see countless icons everywhere.
The word "defaults" recalls another bug that hits me: every time I update the program I lose my personal preferences. Unfortunately I never remember how to set them. There are too many options, most of them undesired and not understood, they are located in many places, some of them are document-specific, others are application-global. A possible cause of my disorientation can be the fact that I don't like Windows. In my former group, most of the members, like me, came from the Macintosh and SwaN-MR. The year before our company forced us to switch to Windows, it had happened that a young researcher had joined our group and she found similar difficulties, because she was switching from Windows/Mestre-C to Mac/SwaN-MR. There were two fundamental differences, though:
- I can show what is missing into Mestre-C, she didn't explain what was wrong with SwaN-MR.
- After one year she felt at ease with SwaN-MR and was productive with it. The more I use Mestre-C, the less I like it.
Can you see why I am curious to see MestreNova? Don't believe that the rest of Mestre-C was OK. Remember that there is a space for comments here below, so you can correct me if I am wrong, or add your personal experience. Regarding mine, it was limited to what I call "routine processing". Nor is it simple to learn the rest. Every command adds its own peculiar interface. You have to remember what a right-clicks performs under each circumstance. Even when the interface is the same, a surprise can arrive like a punch in the stomach. Phase correction, for example, has a unified interface in 1D and 2D. In the latter case I can't specify a pivot point. Why? I ignore it. I only know that it's hard to correct the phase without a pivot. To continue the torture, in 2D, you also have plenty of confusing colors.
Of course I have always expressed, along the years, my criticism to the author, detailing it more than today. The usual answer was the unarguable: "Customers like it in this way". Another common, and not acceptable, answer was: "The bug will disappear with MestreNova". The most important answer, although a silent one, is MestreNova itself: the fact that they have rewritten it all sounds like a condemnation to death for Mestre-C. Let us remember the good qualities of the defunct. The manual, a little outdated, is readable and helpful. The printed output is OK. The job, eventually, is done. Without enjoyment, yet done. For some users it can also be great to know that there are so many advanced options in store.
I am curious: will everything change with the advent of MestreNova? If the answer is yes, will the users be happy to change their habits, to learn new commands, to adopt a new mentality? Most of all: will the heavy approach survive?
Sunday, 12 November 2006
Trip
A complete tour of iNMR would be too lengthy. Today we'll only discover the latest addition, the deconvolution module. Nothing to write home about, if you are already acquainted with iNMR. Linux users have no possibility to see the program in action. This trip is dedicated to them.
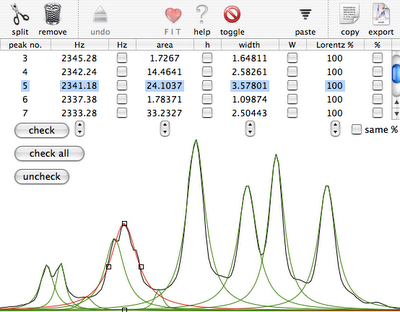
You start selecting a region of a 1D spectrum, already completely processed. The command "Deconvolution" creates the window shown in the first picture. In normal circumstances we'd enlarge the window as much as we like. For the sake of readability of this blog, we keep it small. You can see the experimental spectrum in black and the peaks already fitted (in a very approximative way) with green lorentzian shapes. The selected peak, in red, has a shoulder on the right, but only in the experimental spectrum. To create it in the simulated spectrum too, first we split the peak in two. The leftmost icon performs this operation on the selected peak.
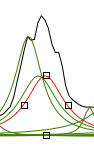
The second picture shows the two halves, red on the right. green on the left. By dragging the square black handles, we can manually fit the peak and the shoulder (see below).
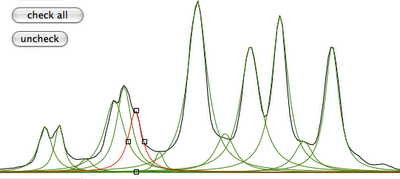
It's now necessary to observe our data differently. With the command "toggle", the green shape corresponds to the sum of the peaks (i.e. the simulated spectrum), while the red line corresponds to the difference (experimental - simulated).
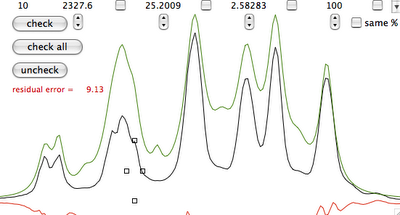
Manual fitting is out of question. Before performing automatic fitting, we have to specify which parameter should be changed. In this case, a click on the button "check all" selects all parameters for adjustment. Another click on the heart icon and we arrive at the last picture.
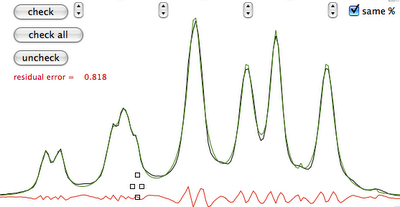
Summary: iNMR specifies the initial parameters, but you can add or remove peaks, shift and reshape them. You have a table at the top if you prefer a numerical input. In this way you can specify: position, shape (lorentzian, gaussian or anything in between), area and width for each line. Graphically you specify: position, height and width. You can mix manual and automatic fitting, the former extended to all parameters or only to selected ones. You can optionally specify that all peaks are described by the same function.
Eventually you get a good estimate of all parameters (e.g.: area, position...) for each peak, even the hidden ones.

You start selecting a region of a 1D spectrum, already completely processed. The command "Deconvolution" creates the window shown in the first picture. In normal circumstances we'd enlarge the window as much as we like. For the sake of readability of this blog, we keep it small. You can see the experimental spectrum in black and the peaks already fitted (in a very approximative way) with green lorentzian shapes. The selected peak, in red, has a shoulder on the right, but only in the experimental spectrum. To create it in the simulated spectrum too, first we split the peak in two. The leftmost icon performs this operation on the selected peak.

The second picture shows the two halves, red on the right. green on the left. By dragging the square black handles, we can manually fit the peak and the shoulder (see below).

It's now necessary to observe our data differently. With the command "toggle", the green shape corresponds to the sum of the peaks (i.e. the simulated spectrum), while the red line corresponds to the difference (experimental - simulated).

Manual fitting is out of question. Before performing automatic fitting, we have to specify which parameter should be changed. In this case, a click on the button "check all" selects all parameters for adjustment. Another click on the heart icon and we arrive at the last picture.

Summary: iNMR specifies the initial parameters, but you can add or remove peaks, shift and reshape them. You have a table at the top if you prefer a numerical input. In this way you can specify: position, shape (lorentzian, gaussian or anything in between), area and width for each line. Graphically you specify: position, height and width. You can mix manual and automatic fitting, the former extended to all parameters or only to selected ones. You can optionally specify that all peaks are described by the same function.
Eventually you get a good estimate of all parameters (e.g.: area, position...) for each peak, even the hidden ones.
Saturday, 11 November 2006
Welcome
When I was convinced that nobody was reading this blog, I have found a pair of comments posted by the product manager of NMR Suite. I am glad to give space to my colleagues and to see them write in plain language, like Jack did. I invite other visitors to read the comments and to download the demo of the program. I hope to find the time in December. In this case I will also publish my detailed review of the latest version. Certainly I am not into metabolomics and I will mainly examine the processing module. For the moment being, I thank Jack because his contribution has enriched this blog.
There will soon be another product to uncover, namely MestreNova. Stay tuned.
There will soon be another product to uncover, namely MestreNova. Stay tuned.
Wednesday, 8 November 2006
Fables
Yesterday Chenomx announced the availability of version 4.6 of their flagship product "NMR Suite". I don't know if I can consider this product a true processing software, albeit the name. They appear very determined, however, and I believe that their NMR suite will eventually process spectra. I can't include it into my price comparison list because they don't publish the price. I believe a single licence is $35,000 (commercial) or $8,750 (academic). Just like ACDlabs, they come from Canada (the land of NMR software?) and have apparently materialized from nothing. Please don't say that ACDlabs is different because it's half Russian, because the name of Chenomx's representative is Alex Cherniavsky.
The whole official story is contained into a single page. In April 2004 "Varian, Inc. has made a strategic equity investment in Chenomx, Inc., a privately held company that is a leader in the rapidly growing field of metabolic profiling" which means:
The next two titles try to catch my eyes with "$18m Genome Canada Grant" and "$2.6m Magnetic Resonance Diagnostics Centre". You know, if you have a grant you can't use freeware nor shareware, you have to spend it all. Under such pressure, the quality of what you buy is not relevant.
Fast forward to one year ago, when NMR Suite finally appears, with version 4.0. The leader of the market can't start from 1, like you and me, he starts directly from 4. We arrive in April when, take a long breath, "Chenomx, Inc., a leading provider of software and compound libraries used to identify and quantify metabolites in NMR spectra, and Umetrics AB, a worldwide leader in multivariate analysis and modeling software, are pleased to announce a partnership to provide a comprehensive metabolomics solution to researchers worldwide". Anybody can be a leader, that's the beauty of the web.
In June we saw version 4.5 and, yesterday, the revolutionary 4.6, which: can now import raw Bruker spectra (what have you been doing up to now?), and will now look and feel more like a native Windows application (sorry you have renounced to your minimalist look, it was your ace in the hole).
I have downloaded and tried the previous version (4.5). For simplicity I limited myself to open the only example included. It was terribly out-of-phase. Though I have matured a decennial experience in phasing with 8 different programs, I could not correct it neither manually nor automatically. The slider reached the end of the run and there was nothing else to do but to ask for help, via e-mail, to Chenomx itself:
"Just one question. I have found a single sample file and I wasn't able to correct the phase. What's the value for the first order phase correction?" And I expected a numerical value. Remember that it was their spectrum, not mine! They replied that the answer is inside the tutorial. It was not, but the picture was clear: they are not ready yet. Maybe in 2007.
The whole official story is contained into a single page. In April 2004 "Varian, Inc. has made a strategic equity investment in Chenomx, Inc., a privately held company that is a leader in the rapidly growing field of metabolic profiling" which means:
- They were already the leaders of the field before having a single product.
- Varian smartly concluded that, if themselves were able to make money with VNMRJ, the software market is where you can become rich.
The next two titles try to catch my eyes with "$18m Genome Canada Grant" and "$2.6m Magnetic Resonance Diagnostics Centre". You know, if you have a grant you can't use freeware nor shareware, you have to spend it all. Under such pressure, the quality of what you buy is not relevant.
Fast forward to one year ago, when NMR Suite finally appears, with version 4.0. The leader of the market can't start from 1, like you and me, he starts directly from 4. We arrive in April when, take a long breath, "Chenomx, Inc., a leading provider of software and compound libraries used to identify and quantify metabolites in NMR spectra, and Umetrics AB, a worldwide leader in multivariate analysis and modeling software, are pleased to announce a partnership to provide a comprehensive metabolomics solution to researchers worldwide". Anybody can be a leader, that's the beauty of the web.
In June we saw version 4.5 and, yesterday, the revolutionary 4.6, which: can now import raw Bruker spectra (what have you been doing up to now?), and will now look and feel more like a native Windows application (sorry you have renounced to your minimalist look, it was your ace in the hole).
I have downloaded and tried the previous version (4.5). For simplicity I limited myself to open the only example included. It was terribly out-of-phase. Though I have matured a decennial experience in phasing with 8 different programs, I could not correct it neither manually nor automatically. The slider reached the end of the run and there was nothing else to do but to ask for help, via e-mail, to Chenomx itself:
"Just one question. I have found a single sample file and I wasn't able to correct the phase. What's the value for the first order phase correction?" And I expected a numerical value. Remember that it was their spectrum, not mine! They replied that the answer is inside the tutorial. It was not, but the picture was clear: they are not ready yet. Maybe in 2007.
Tuesday, 7 November 2006
Tutorial
This tutorial is dedicated to Windows users. Visit SDBD with the following link or, if it doesn't work, search the compound no. 548 (o-dichlorobenzene, C6 H4 Cl2). You find the following table:
Parameter ppm Hz
D(A) 7.366
D(B) 7.110
J(A,A') 0.32
J(A,B) 8.04
J(A,B') 1.54
J(A',B) 1.54
J(A',B') 8.04
J(B,B') 7.45
CASTELLANO,S. & KOSTELNIK,R. TETRAHEDRON LETT. 1967, 5211
Just because you don't have Mac OS X, there is no reason to download iNMR. If you could run it, you'd create a simulated spectrum with Cmd-N.
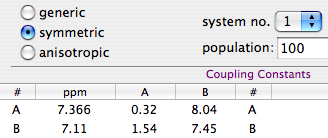
First Thing First: select the option "symmetric". This allows us to only specify two of the four hydrogens. Copy the ODCB values as shown in the picture, then click OK. Otherwise, download the corresponding file: odcb.spins. Now you can generate synthetic spectra at any observe frequency. You can use the keyboard or the interactive control (for continuous variation). The pictures show the results at 60 and 300 MHz.
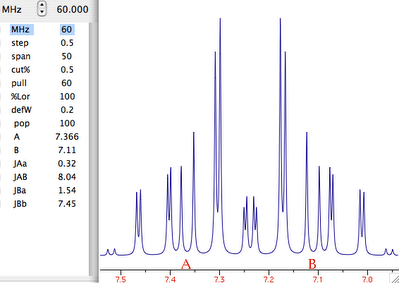
You can also play with the chemical shifts and the coupling constants using the same mechanism.
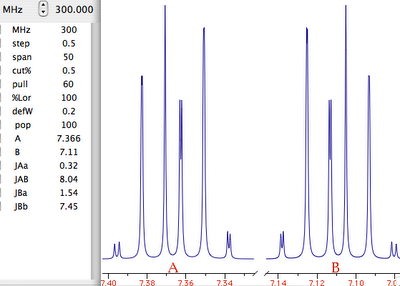
Parameter ppm Hz
D(A) 7.366
D(B) 7.110
J(A,A') 0.32
J(A,B) 8.04
J(A,B') 1.54
J(A',B) 1.54
J(A',B') 8.04
J(B,B') 7.45
CASTELLANO,S. & KOSTELNIK,R. TETRAHEDRON LETT. 1967, 5211
Just because you don't have Mac OS X, there is no reason to download iNMR. If you could run it, you'd create a simulated spectrum with Cmd-N.

First Thing First: select the option "symmetric". This allows us to only specify two of the four hydrogens. Copy the ODCB values as shown in the picture, then click OK. Otherwise, download the corresponding file: odcb.spins. Now you can generate synthetic spectra at any observe frequency. You can use the keyboard or the interactive control (for continuous variation). The pictures show the results at 60 and 300 MHz.

You can also play with the chemical shifts and the coupling constants using the same mechanism.

Empirical Approach
How to get a flat base-plane in multidimensional NMR? Many years ago they told me to multiply the first point of the FID by 0.5. Many colleagues know it. You can find the rationale in the literature. I too could find it, just was too lazy for studying. To justify my laziness my arguments were:
Another questionable practice is the baseline correction performed on the transformed increments, soon after the FT along the direct dimension. It must be automatic (normally you don't even see the imaginary part) and the spectrum is still complex. How can an algorithm find a region of pure baseline into a crowded dispersion spectrum? How can you trust into it, without a visual feed-back? You don't have this problems with first point pre-multiplication. It's so pure, you know. Like homeopathy, it can't hurt. This is why I used the latter, like everybody else. Mestre-C still applies it by default on all spectra (1D included, I mean).
With the Mac Quadra, running at 25 MHz, it was possible to transform a matrix in less than two minutes. Finally I could optimize my processing parameters by trial and error. I pre-multiplied by 0, 0.25, 0.5, 0.75 and by 1.25. Judging from the final result (after complete processing, base-plane correction included), my first spectrum preferred no pre-multiplication. My second spectrum, a week later, preferred no pre-multiplication. The third spectrum did just the same. I declared my experimentation period terminated. I haven't kept the results with me. From that day I simply stopped pre-multiplying and my spectra were not so bad. Since the introduction of the digital filter by Bruker, the first point is zero or near-zero, so in that case you can even multiply by 10, if you are in the mood.
- The literature is very old. If what they say is right, manufacturers, who read it before me, are already accounting for it and the acquisition software already pre-multiplies the first point.
- OK, the first increment of a 2D doesn't look pre-multiplied. But my spectrum still needs a baseline correction after FT, so it makes no difference.
- The literature predates phase-sensitive 2Ds, predates 2D baseline corrections, predates fast computers. Probably it's not valid for today's spectra. Probably they had to avoid baseline correction, because their computers were too slow, and invented this poor man's alternative.
Another questionable practice is the baseline correction performed on the transformed increments, soon after the FT along the direct dimension. It must be automatic (normally you don't even see the imaginary part) and the spectrum is still complex. How can an algorithm find a region of pure baseline into a crowded dispersion spectrum? How can you trust into it, without a visual feed-back? You don't have this problems with first point pre-multiplication. It's so pure, you know. Like homeopathy, it can't hurt. This is why I used the latter, like everybody else. Mestre-C still applies it by default on all spectra (1D included, I mean).
With the Mac Quadra, running at 25 MHz, it was possible to transform a matrix in less than two minutes. Finally I could optimize my processing parameters by trial and error. I pre-multiplied by 0, 0.25, 0.5, 0.75 and by 1.25. Judging from the final result (after complete processing, base-plane correction included), my first spectrum preferred no pre-multiplication. My second spectrum, a week later, preferred no pre-multiplication. The third spectrum did just the same. I declared my experimentation period terminated. I haven't kept the results with me. From that day I simply stopped pre-multiplying and my spectra were not so bad. Since the introduction of the digital filter by Bruker, the first point is zero or near-zero, so in that case you can even multiply by 10, if you are in the mood.
Pipe mentality
An helpful wiki/tutorial of NMRPipe serves as a review as well. A few excerpts will give you a picture of the proteinist mentality (to be compared with the CW mentality).
.....
It is important to realize that processing this data requires you to frequently exit and enter NMRPipe to work with text editors and invoke commands on command lines.
.....
then left-click on “Execute” to apply a cosine-bell (90°-shifted sine-bell) window function. [...] Write down the parameters used for this function because you’ll need to enter them into your script later.
.....
Once you are satisfied with the phasing of your spectrum, write down the P0 and P1 values so you can use them in your processing script.
.....
The script shown above should produce a good spectrum under most circumstances.
.....
Edit parameters for processing the indirect dimension. You don’t get to see a 1D spectrum to help you alter these parameters. In the NMRPipe procedure, you just process the 2D spectrum as a whole, read it into the GUI, make observations and phase adjustments in the GUI, then return to this script, alter it, and reprocess the spectrum.
Processing a 2D spectrum with NMRPipe is a ritual. If you are going to spend 2 weeks (or more) to assign your spectrum, why not spending a whole morning to process it? In iNMR you get the same results with:
From the tutorial I get the impression that the majority of NMRPipe users spend their time in copying the same script with the same default commands in all their folders, to get the same standard processing (see text in bold). What's the point? How did they arrive there? NMRPipe started as a program for programmers. Modifying the script the user can save the half-processed spectrum at any time, apply on it another program (likely written by himself), then return to NMRPipe. In a different scenario, the user can just learn to write the scripts (a basic form of programming) and experiment with custom work-flows. NMRPipe is a useful tool and it's simple to understand. Being it free, it's also popular. When it appeared, it obviously looked more advanced than now.
The "proteinist mentality" thinks: 1D NMR belongs to the past; NMR and "Protein NMR" are the same thing; I'll write my own plug-ins for whichever program I am gonna use.
I am exaggerating! I can't say I know a single person who thinks in this way. Still I have often heard the observation that there is ample choice of software, both commercial and free, and no space for a new product. When you carefully compare two programs, you can actually notice so vast differences in approach to justify their simultaneous existence.
I don't know if the popularity of NMRPipe is still growing or decreasing. For years, I have been visiting weekly a "Large Research Infrastructure"where 90% or more of the researches preferred XWin-NMR and TopSpin, and my impression is that the latter peaked at the 100%. Is NMR processing so mature that the industrial product has superseded the research tool? Or is Bruker investing, at last, big money in software? Or, more simply, people had enough of command line tools?
.....
It is important to realize that processing this data requires you to frequently exit and enter NMRPipe to work with text editors and invoke commands on command lines.
.....
then left-click on “Execute” to apply a cosine-bell (90°-shifted sine-bell) window function. [...] Write down the parameters used for this function because you’ll need to enter them into your script later.
.....
Once you are satisfied with the phasing of your spectrum, write down the P0 and P1 values so you can use them in your processing script.
.....
The script shown above should produce a good spectrum under most circumstances.
.....
Edit parameters for processing the indirect dimension. You don’t get to see a 1D spectrum to help you alter these parameters. In the NMRPipe procedure, you just process the 2D spectrum as a whole, read it into the GUI, make observations and phase adjustments in the GUI, then return to this script, alter it, and reprocess the spectrum.
Processing a 2D spectrum with NMRPipe is a ritual. If you are going to spend 2 weeks (or more) to assign your spectrum, why not spending a whole morning to process it? In iNMR you get the same results with:
- two clicks on the "gears" icon;
- one click on the manual phase correction icon, followed by the interactive adjustment of the whole matrix;
- one click on the automatic baseline correction icon.
From the tutorial I get the impression that the majority of NMRPipe users spend their time in copying the same script with the same default commands in all their folders, to get the same standard processing (see text in bold). What's the point? How did they arrive there? NMRPipe started as a program for programmers. Modifying the script the user can save the half-processed spectrum at any time, apply on it another program (likely written by himself), then return to NMRPipe. In a different scenario, the user can just learn to write the scripts (a basic form of programming) and experiment with custom work-flows. NMRPipe is a useful tool and it's simple to understand. Being it free, it's also popular. When it appeared, it obviously looked more advanced than now.
The "proteinist mentality" thinks: 1D NMR belongs to the past; NMR and "Protein NMR" are the same thing; I'll write my own plug-ins for whichever program I am gonna use.
I am exaggerating! I can't say I know a single person who thinks in this way. Still I have often heard the observation that there is ample choice of software, both commercial and free, and no space for a new product. When you carefully compare two programs, you can actually notice so vast differences in approach to justify their simultaneous existence.
I don't know if the popularity of NMRPipe is still growing or decreasing. For years, I have been visiting weekly a "Large Research Infrastructure"where 90% or more of the researches preferred XWin-NMR and TopSpin, and my impression is that the latter peaked at the 100%. Is NMR processing so mature that the industrial product has superseded the research tool? Or is Bruker investing, at last, big money in software? Or, more simply, people had enough of command line tools?
Monday, 6 November 2006
When I did not create a new file format
What follows is the letter that, in February, explained to the users the iNMR storing strategy.
Amid so many news, now it's finally the time for me to illustrate the great innovation that version 1 brings to the field of NMR software. Up to now, it was customary for any new program to create its own new format for files, or (in exceptional cases) to adopt only one of the existing formats. This year I finally materialized an old idea of mine, namely to create a new application without creating a new format and potentially accepting any format, and whose files can be read by already existing applications. It works like this:
The overall concept seems easy, but it required a lot of programming effort. This is the reason why no other program (as far as I know) adopted a similar solution. I do hope you will appreciate the advantages of it. Consider the common example of someone having used SwaN-MR for a decade and now possessing a large library of spectra in that format (whose description has been publicly available for years). Today only Mestre-C and iNMR can read those files (I don't know why others don't). If you, instead, start now to create a library of iNMR spectra, you can (from day one) open the spectra with any software. Furthermore, iNMR doesn't need to read and convert all the acquisition parameters, because they are preserved in their original files. Other parameters (those specific to iNMR) can already be read by two ubiquitous Apple applications and, being in standard XML format, any programmer can manipulate them.
Amid so many news, now it's finally the time for me to illustrate the great innovation that version 1 brings to the field of NMR software. Up to now, it was customary for any new program to create its own new format for files, or (in exceptional cases) to adopt only one of the existing formats. This year I finally materialized an old idea of mine, namely to create a new application without creating a new format and potentially accepting any format, and whose files can be read by already existing applications. It works like this:
- Every time you open a spectrum, iNMR restarts from the original spectrometer files. In this way users are enforced to the safe practice of storing raw experimental data. Too many times I have seen unaware researchers store processed data and throwing away the FID, on the erroneous assumption that the FID is useless. Actually it's the processed data that is often useless (unless we are talking about 3D spectra).
- Your processing work is saved as parameters into a separate, easily recognizable file, and iNMR will reprocess everything automatically for you when you are returning to a spectrum.
- The iNMR files can be read and written with an Apple application that all of you already have installed, called "Property List Editor". This is why I say I have not created a new file format. I do suggest you to open a file in this way, to have an idea of how everything works. Be careful not to edit the file, because you can easily make iNMR crash.
- You can also open the iNMR files with TextEdit, TextWrangler or an equivalent application. From the first lines you will see that they are also XML files. I am sure you have already heard about it. XML is a great solution for storing data and it's likely that in a near future every application will be adopting it (many already do). It's certainly light years ahead of JCAMP-DX.
- Having adopted all the latest technologies, like Quartz for graphics and XML for files, iNMR will be compatible with future versions of the Mac OS for many years to come.
- You can already open iNMR spectra with other existing software. It will not recognize the XML files, but it will recognize the original spectrometer files, because they are never modified or renamed.
The overall concept seems easy, but it required a lot of programming effort. This is the reason why no other program (as far as I know) adopted a similar solution. I do hope you will appreciate the advantages of it. Consider the common example of someone having used SwaN-MR for a decade and now possessing a large library of spectra in that format (whose description has been publicly available for years). Today only Mestre-C and iNMR can read those files (I don't know why others don't). If you, instead, start now to create a library of iNMR spectra, you can (from day one) open the spectra with any software. Furthermore, iNMR doesn't need to read and convert all the acquisition parameters, because they are preserved in their original files. Other parameters (those specific to iNMR) can already be read by two ubiquitous Apple applications and, being in standard XML format, any programmer can manipulate them.
JCAMP-DX criticism
The JCAMP-DX protocol is a pair of articles, republished as scanned images. This fact alone, compared to the XML protocol says it all: with no money, you cannot be elegant and good-looking. The fact that the protocol looks so old, combined with the sentence: The commitment of three leading NMR spectrometer manufacturers at the Sixth INUM Conference in Italy 1990 to implement the standard when finished... makes me feel like an historical character, like if I had been at the Yalta Conference. I was indeed a registered attendee of the 6th INUM Conference, and I cannot forget it because it was my first time in the lovely Stresa. I was a completely passive attendee, of course, and it was also the first time I heard about the JCAMP standard, which seemed to me a wonderful idea. I hoped for a universal format for all spectra and easy connection of all spectrometers to PCs. The second hope has materialized after many years. The first hope soon faded away and had no reason to exist in the first place. Now that I am a player in the field I know that it's plainly impossible nor desirable. Then I was young and hopeful. The mere fact to be there, as the representative of a multinational corporation, was an accomplishment. Now it seems a dream. I really wonder if it was all true. Enough for the personal considerations. Back to the file format.
If you read the article you can't understand the protocol, because it is described elsewhere, in the infrared section. This is only the first fault: the protocol had to be contained into a single document. Let's count them all.
Creating an NMR standard is difficult and unpleasant. It was a dirty job and somebody did it and it even works (some times). I am not able to do any better. The good news is that nobody cares, therefore: why should we care?
If you read the article you can't understand the protocol, because it is described elsewhere, in the infrared section. This is only the first fault: the protocol had to be contained into a single document. Let's count them all.
- The lack of a single reference text. Each new version should have completely substituted the old ones (like the pharmacopeia).
- Definitions are not complete. A sentence like: "FIDs will be handled differently depending on whether they originate from a spectrometer with a single detector and analog-to-digital converter (ADC) or with twin detectors and a single ADC." is clear but doesn't really help. How is the FIDs to be handled? How digital instruments enter into this picture?
- It was probably OK for IR, but when extended to 1D NMR it was not so great. Extension to 2D is questionable. They had to go from the general to the particular, but they went the other way.
- Let's not forget that today we have XML.
- JCAMP-DX, when it works, is simply a mean for data exchange, not for archival. It is not considered by regulatory agencies.
- It allows the creation of too many flavors of files.
- There is no validator.
- Existing programs both write and read (without warnings) files that are not completely compliant with the published guidelines.
- Numbers are rounded so, whatever the article says, the conversion into JCAMP-DX generates a small loss of information. This loss is highly variable.
- The only thing that the existing file formats had in common, is that they only store the intensity values. JCAMP-DX is the only NMR file format that stores the abscissa values.
- It is not popular.
- They forgot that NMR is not limited to chemicals. Spectra of living beings are not considered.
Creating an NMR standard is difficult and unpleasant. It was a dirty job and somebody did it and it even works (some times). I am not able to do any better. The good news is that nobody cares, therefore: why should we care?
Integral labels
Q: Can you set the integrals so they read vertical instead of horizontal?
A: The integrals are written horizontally. You also wrote your e-mail horizontally. Why do you want to write vertically? It's more difficult to read. Your eyes will soon get tired. There are many tricks to avoid overcrowding of integrals:
A: The integrals are written horizontally. You also wrote your e-mail horizontally. Why do you want to write vertically? It's more difficult to read. Your eyes will soon get tired. There are many tricks to avoid overcrowding of integrals:
- reduce the number of digits.
- set 1 proton = 100 integral units and you can do without decimal digits.
- enlarge some integrals to the right or to the left: the number will be displaced accordingly.
- change the font.
- use the cutter tool to remove non-interesting regions.
- print on larger sheets of paper (we in Europe have the A3 format).
- you can merge two integral regions; you will only read the sum, yet it's usually trivial to estimate by eye the contributions of each peak.
An historical picture
The picture in this page is a good example of the power of SwaN-MR. It demonstrates that the peak on the left should be assigned to residue no. 3 (Hyp) instead of no. 8 (Oic) as erroneously published on J. Am. Chem. Soc. 1994, 116, 7532-7540.
One may object that I unmasked the error because I acquired the spectrum again and not because of SwaN-MR. If this is true, it is also true that SwaN-MR gave me a simple and elegant way to demonstrate my thesis. By the way: generating the above gif image has also been incredibly simple with SwaN-MR. The whole work was done on a Quadra 610 upgraded to the PowerPC.
Dear Giuseppe Balacco,
an interesting picture located at http://qobrue.usc.es/jsgroup/Swan/ Found this more accidentally. Nevertheless this isn't correct in the manner described. Of course you are unfortunately right concerning the misassignment (as stated few years ago). But this was no problem concerning the spectra quality but a change of the substance between the acquisition of the spectra for assignment purposes and the NOESY spectrum.
Best regards
Rainer Haessner
Dear Rainer Haessner, I wish you can tell the whole story on your own blog, maybe when Kessler's reaction can no more affect your career.
When I did not invent monochromatic spectra
Another example of how Varian software influenced me is the number of colors I have used to draw the contour levels of a 2D spectrum. Not just the Varian software, but all the programs that I have met, use a different color for each level. (20 levels = 20 colors). I limited myself to only 6 colors, but I wasn't satisfied. My spectra were difficult to read. Other users also complained for the paucity of colors. In my adolescence I had been extremely fond of maps and topography. I really studied it a lot, so I was used to contour levels well before the introduction of color printers and color monitors (CGA, EGA, VGA...). I remembered that old maps were black & white, and newer maps had color, and the latter were more readable BUT they didn't mix colors. Green was employed for vegetation, blue for water, brownish for the contour levels, black for names, etc... I tried to use a single color for positive levels and another one for negative levels. Suddenly, all became beautiful, evident, mind-relaxing. Three subjective reactions, I agree, but there was a good reason for a generalization. Don't you think that topography is a science? That they have arrived at the same conclusion after studying? They had already found the answer long ago.
At the same time I began overlapping my spectra. Either on screen or on paper, I always displayed the COSY (red), NOESY (blue/cyan) and TOCSY (black) together (not stacked: _overlapped_ ), obviously with 1D projections and assignment of each cross-peak. The total was more readable that the single parts alone! This was obviously impossible conserving the popular habit of changing the color at each level. I knew I was right. As it happens, people were not yet convinced. After being conditioned by miseducative software, they wanted to remain harlequins forever.
It all changed when I began drawing contour maps with anti-aliasing and double-buffering. As soon as I saw the effect on a 20" LCD monitor, with me playing the computer keyboard and the display responsive in real time, I murmured: "Questo e' l'NMR come nessuno l'ha mai provato" which means: "This is the ultimate NMR experience". A few months later I uploaded iNMR and it has been downloaded many thousands of times. People have asked for every sort of extension but nobody has ever asked for more colors.
At the same time I began overlapping my spectra. Either on screen or on paper, I always displayed the COSY (red), NOESY (blue/cyan) and TOCSY (black) together (not stacked: _overlapped_ ), obviously with 1D projections and assignment of each cross-peak. The total was more readable that the single parts alone! This was obviously impossible conserving the popular habit of changing the color at each level. I knew I was right. As it happens, people were not yet convinced. After being conditioned by miseducative software, they wanted to remain harlequins forever.
It all changed when I began drawing contour maps with anti-aliasing and double-buffering. As soon as I saw the effect on a 20" LCD monitor, with me playing the computer keyboard and the display responsive in real time, I murmured: "Questo e' l'NMR come nessuno l'ha mai provato" which means: "This is the ultimate NMR experience". A few months later I uploaded iNMR and it has been downloaded many thousands of times. People have asked for every sort of extension but nobody has ever asked for more colors.
Discussion
I have extracted the following dialog from the vault where it was hidden and forgotten. Before you read it, let me establish the undisputed facts:
NMR Guy 02-09-2006, 01:06 AM
A version of the processing module for VNMRj is available for OS X. To be honest, if you are serious about NMR, get the Varian VNMRj OS X version... or x-windows in to VNMR/VNMRj (Varian) running on Solaris or Linux, or TopSpin (Bruker) running on Linux. BTW Not sure what the deal with iNMR and SwaN-MR is. Looks like SwaN-MR (freeware/registerware) has been taken down completely and replaced with this software, which is not all that cheap.
bazzler 02-10-2006, 05:56 AM
I am a mac-using organic chemist. Until now, there has never been an app for nmr reprocessing app for OSX that was powerful and userfriendly. iNMR is intuitive to use and pleasing to the eye, very much in the way as one would expect from an Apple application. It performs many (including all of the important and critical) functions of the existing nmr reproc. solutions, and gives the output i want with the minimum of effort. I cannot recommend highly enough this elegant and powerful program to anyone involved with NMR, regardless of their computing background. I am not alone in my praise either, their are several mac - chemists in the department where i work , and like me they were all blown away with the excellence of this software. The mac is somewhat of a forsaken child in the world of chemistry, particularly with 3rd party apps; the author should be commended for daring to invest his time and effort with this gem of a program. If youre an organic chemist, give it a go and spread the word; you wont be disappointed.
- The last version of SwaN-MR shipped in 2001, while iNMR 1.0 shipped in 2006. There is no connection between the two programs.
- iNMR has been written from scratch, just like SwaN-MR. Like "La Traviata" and "Aida" they have been written by the same hand, so you can recognize a common style in the two opuses. Style is a positive quality.
- SwaN-MR has not been replaced. It still works perfectly, as ever since 1994.
- "NMR Guy" compares iNMR with VNMRJ and TopSpin, and says that the former is not cheap. At the price of a single license of VNMRJ you can buy 12 copies of iNMR. At the price of a single license of TopSpin you can buy 30 copies of iNMR. I am speaking of the latest version (1.5), but there is also version 0.7, offered at 50 euro.
- The nearly 300 iNMR users are all enthusiast about it, for the simple reason that they could evaluate it before buying. Most of the VNMRJ and TopSpin users had no choice, because the programs come with the respective instruments.
- There must be a reason why somebody, already owning VNMRJ or TopSpin, invests a few bucks in iNMR. Probably something related to quality...
NMR Guy 02-09-2006, 01:06 AM
A version of the processing module for VNMRj is available for OS X. To be honest, if you are serious about NMR, get the Varian VNMRj OS X version... or x-windows in to VNMR/VNMRj (Varian) running on Solaris or Linux, or TopSpin (Bruker) running on Linux. BTW Not sure what the deal with iNMR and SwaN-MR is. Looks like SwaN-MR (freeware/registerware) has been taken down completely and replaced with this software, which is not all that cheap.
bazzler 02-10-2006, 05:56 AM
I am a mac-using organic chemist. Until now, there has never been an app for nmr reprocessing app for OSX that was powerful and userfriendly. iNMR is intuitive to use and pleasing to the eye, very much in the way as one would expect from an Apple application. It performs many (including all of the important and critical) functions of the existing nmr reproc. solutions, and gives the output i want with the minimum of effort. I cannot recommend highly enough this elegant and powerful program to anyone involved with NMR, regardless of their computing background. I am not alone in my praise either, their are several mac - chemists in the department where i work , and like me they were all blown away with the excellence of this software. The mac is somewhat of a forsaken child in the world of chemistry, particularly with 3rd party apps; the author should be commended for daring to invest his time and effort with this gem of a program. If youre an organic chemist, give it a go and spread the word; you wont be disappointed.
Subscribe to:
Comments (Atom)
